| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
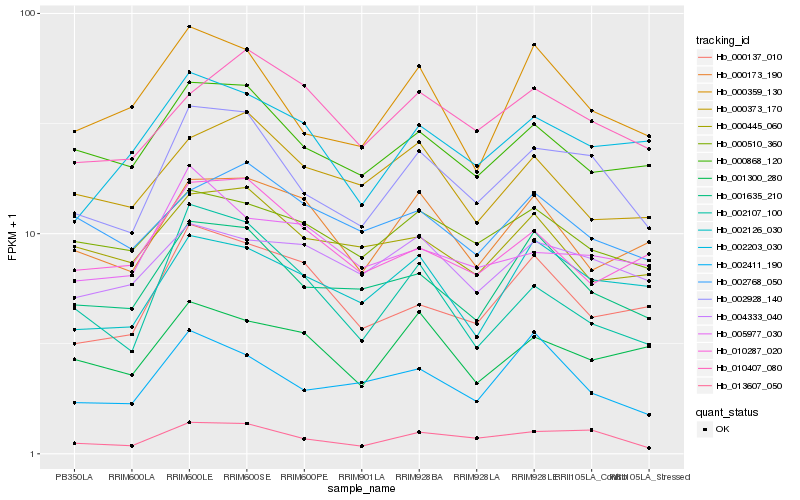
Hb_002928_140 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 2 |
Hb_013607_050 |
0.0614965195 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 2 [Jatropha curcas] |
| 3 |
Hb_001635_210 |
0.0932163014 |
- |
- |
PREDICTED: protein EARLY FLOWERING 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000868_120 |
0.101083536 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 5 |
Hb_000359_130 |
0.1034882113 |
- |
- |
4-hydroxyphenylpyruvate dioxygenase [Hevea brasiliensis] |
| 6 |
Hb_002126_030 |
0.110367868 |
- |
- |
ferrochelatase, putative [Ricinus communis] |
| 7 |
Hb_002203_030 |
0.1159913678 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 8 |
Hb_000510_360 |
0.1183666478 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 9 |
Hb_010407_080 |
0.1194791472 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004333_040 |
0.1218826069 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 11 |
Hb_002107_100 |
0.1245594375 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 12 |
Hb_002411_190 |
0.1246084497 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 13 |
Hb_010287_020 |
0.1252317027 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 14 |
Hb_000373_170 |
0.1265342949 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 15 |
Hb_001300_280 |
0.126792415 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein ucpB [Jatropha curcas] |
| 16 |
Hb_000445_060 |
0.1269889429 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 17 |
Hb_002768_050 |
0.1275301295 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 18 |
Hb_000137_010 |
0.1275667996 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 19 |
Hb_005977_030 |
0.12760096 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 20 |
Hb_000173_190 |
0.1276921735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |