| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
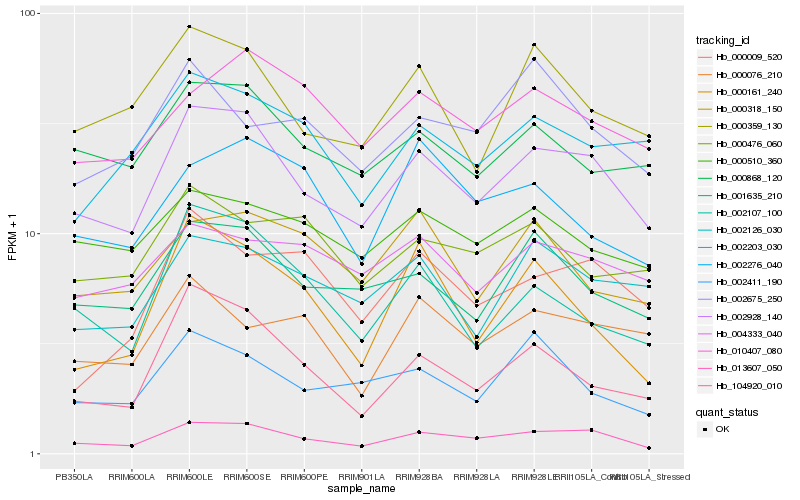
Hb_013607_050 |
0.0 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 2 [Jatropha curcas] |
| 2 |
Hb_002928_140 |
0.0614965195 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001635_210 |
0.136924585 |
- |
- |
PREDICTED: protein EARLY FLOWERING 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010407_080 |
0.1400821146 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000510_360 |
0.1465399544 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 6 |
Hb_002276_040 |
0.1476078771 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 7 |
Hb_000359_130 |
0.1479308709 |
- |
- |
4-hydroxyphenylpyruvate dioxygenase [Hevea brasiliensis] |
| 8 |
Hb_000868_120 |
0.1486310373 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 9 |
Hb_000009_520 |
0.1504366653 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 10 |
Hb_104920_010 |
0.1504540264 |
- |
- |
hypothetical protein VITISV_031859 [Vitis vinifera] |
| 11 |
Hb_002203_030 |
0.1507221002 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 12 |
Hb_000161_240 |
0.1514569521 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002126_030 |
0.1528705552 |
- |
- |
ferrochelatase, putative [Ricinus communis] |
| 14 |
Hb_002411_190 |
0.1528740602 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 15 |
Hb_002675_250 |
0.1529011065 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 16 |
Hb_000076_210 |
0.153482387 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit alpha, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_004333_040 |
0.1536564309 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 18 |
Hb_002107_100 |
0.1544569779 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 19 |
Hb_000476_060 |
0.1574923959 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000318_150 |
0.1586868565 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |