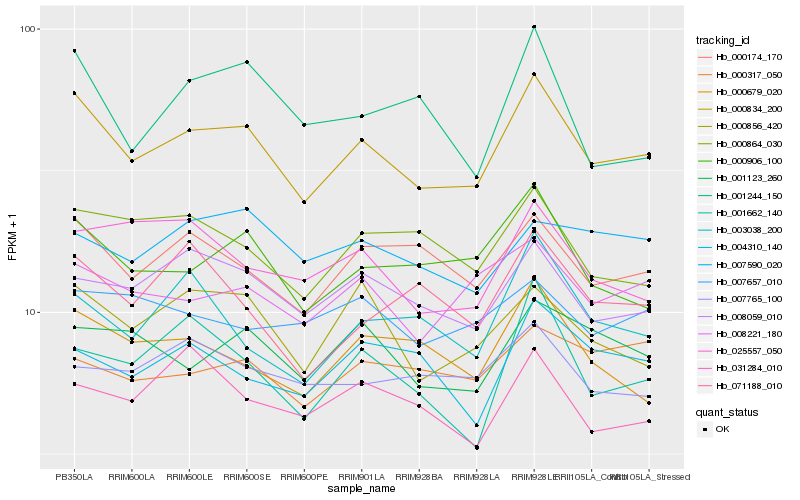
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000834_200 |
0.0 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 2 |
Hb_000174_170 |
0.0772882864 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000906_100 |
0.0783264292 |
- |
- |
PREDICTED: uncharacterized protein LOC105648070 [Jatropha curcas] |
| 4 |
Hb_008059_010 |
0.0790746202 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 5 |
Hb_008221_180 |
0.0821869227 |
- |
- |
PREDICTED: uncharacterized protein LOC105644223 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000856_420 |
0.0824903833 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_000679_020 |
0.087206887 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 8 |
Hb_001123_260 |
0.0874508135 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 9 |
Hb_000864_030 |
0.0890844964 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_025557_050 |
0.0940114354 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 11 |
Hb_001662_140 |
0.0946209213 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004310_140 |
0.0953855387 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_003038_200 |
0.09561303 |
desease resistance |
Gene Name: MMR_HSR1 |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |
| 14 |
Hb_071188_010 |
0.0960131074 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 15 |
Hb_007765_100 |
0.0968119972 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 16 |
Hb_007590_020 |
0.0970490093 |
- |
- |
PREDICTED: protein DAMAGED DNA-BINDING 2 [Jatropha curcas] |
| 17 |
Hb_031284_010 |
0.097185802 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 18 |
Hb_007657_010 |
0.0972600935 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_000317_050 |
0.0972824992 |
- |
- |
formiminotransferase-cyclodeaminase, putative [Ricinus communis] |
| 20 |
Hb_001244_150 |
0.0973428295 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |