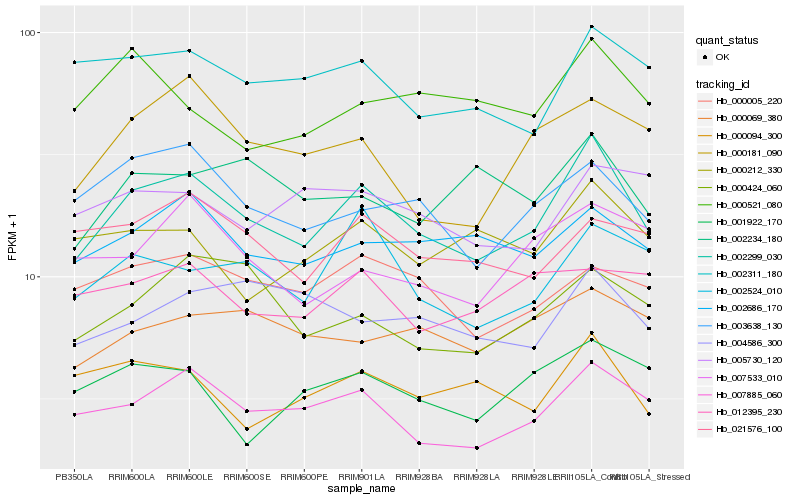
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002299_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007885_060 |
0.0892059675 |
- |
- |
replication factor C 40 kDa family protein [Populus trichocarpa] |
| 3 |
Hb_003638_130 |
0.1103006595 |
- |
- |
PREDICTED: uncharacterized protein LOC105645779 [Jatropha curcas] |
| 4 |
Hb_002686_170 |
0.1126181748 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002524_010 |
0.1133115601 |
- |
- |
hypothetical protein POPTR_0006s20250g [Populus trichocarpa] |
| 6 |
Hb_002311_180 |
0.114925003 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 7 |
Hb_000181_090 |
0.1163284072 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 8 |
Hb_000005_220 |
0.1172180555 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 9 |
Hb_000424_060 |
0.1179867589 |
- |
- |
PREDICTED: myosin-15 [Jatropha curcas] |
| 10 |
Hb_000069_380 |
0.1215288444 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_021576_100 |
0.1224752088 |
- |
- |
Enoyl-CoA hydratase, mitochondrial precursor, putative [Ricinus communis] |
| 12 |
Hb_000094_300 |
0.1233884892 |
- |
- |
hypothetical protein POPTR_0002s26360g [Populus trichocarpa] |
| 13 |
Hb_000212_330 |
0.123599072 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_002234_180 |
0.1239228125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007533_010 |
0.1241450413 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 16 |
Hb_004586_300 |
0.1261492108 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001922_170 |
0.1293870124 |
- |
- |
Plastidic pyruvate kinase beta subunit 1 isoform 2 [Theobroma cacao] |
| 18 |
Hb_012395_230 |
0.1294580104 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 19 |
Hb_000521_080 |
0.1296156134 |
- |
- |
Zinc finger BED domain-containing 4 [Gossypium arboreum] |
| 20 |
Hb_005730_120 |
0.1297880178 |
- |
- |
PREDICTED: protein WHI4 [Jatropha curcas] |