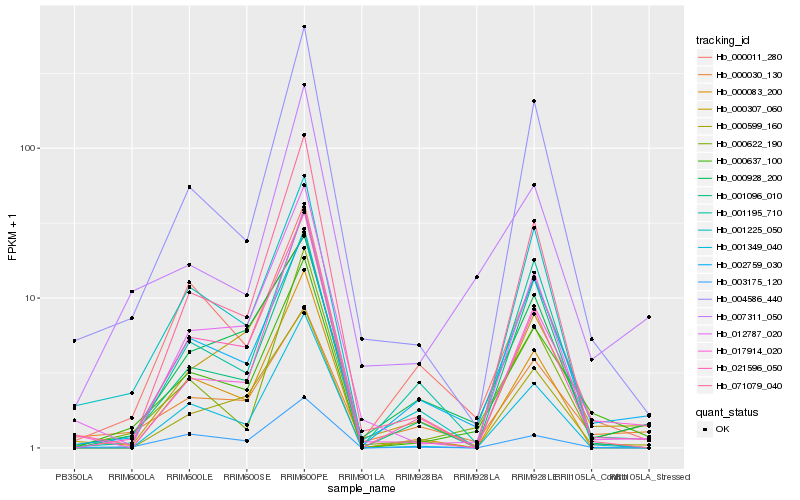
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000637_100 |
0.0 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Jatropha curcas] |
| 2 |
Hb_002759_030 |
0.136779138 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 3 |
Hb_000622_190 |
0.1417874584 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 4 |
Hb_021596_050 |
0.1489872734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004586_440 |
0.152440715 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 6 |
Hb_000030_130 |
0.1598571805 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000599_160 |
0.1612728237 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 8 |
Hb_007311_050 |
0.1665145024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001096_010 |
0.1674704941 |
- |
- |
PREDICTED: uncharacterized protein LOC105650448 [Jatropha curcas] |
| 10 |
Hb_012787_020 |
0.1682362543 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 11 |
Hb_000928_200 |
0.168836495 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 12 |
Hb_003175_120 |
0.1691760039 |
- |
- |
PREDICTED: uncharacterized protein LOC105141472 isoform X1 [Populus euphratica] |
| 13 |
Hb_000307_060 |
0.1706103391 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 14 |
Hb_001225_050 |
0.1713377718 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 15 |
Hb_000011_280 |
0.1735641052 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 16 |
Hb_001349_040 |
0.1741092334 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 17 |
Hb_000083_200 |
0.1741796008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_071079_040 |
0.1744037295 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET3 [Jatropha curcas] |
| 19 |
Hb_001195_710 |
0.1744997645 |
- |
- |
glycosyltransferase family GT8 protein [Populus deltoides] |
| 20 |
Hb_017914_020 |
0.174698031 |
- |
- |
- |