| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
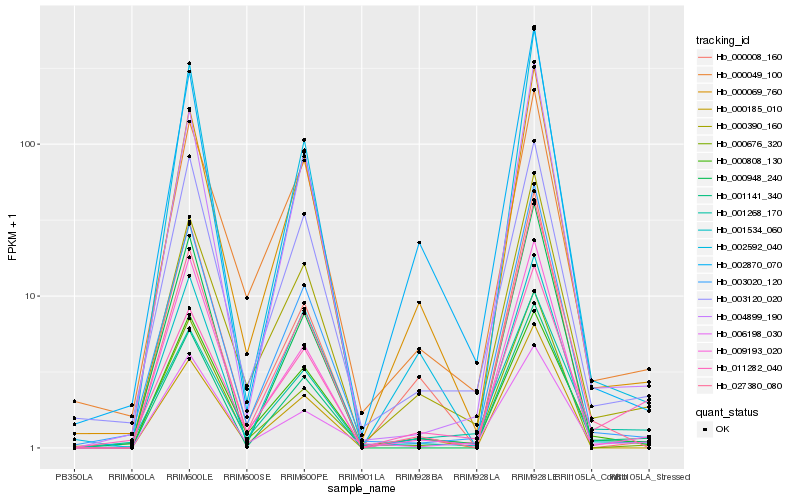
Hb_001141_340 |
0.0 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 2 |
Hb_000069_760 |
0.0823780635 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000390_160 |
0.0858662477 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 4 |
Hb_003120_020 |
0.0949651766 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 13-like [Prunus mume] |
| 5 |
Hb_002592_040 |
0.0973024632 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 6 |
Hb_004899_190 |
0.1024935928 |
- |
- |
RecName: Full=Peroxiredoxin Q, chloroplastic; AltName: Full=Thioredoxin reductase; Flags: Precursor [Populus x jackii] |
| 7 |
Hb_001268_170 |
0.102666677 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 8 |
Hb_003020_120 |
0.1045063113 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 9 |
Hb_009193_020 |
0.108706295 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 10 |
Hb_002870_070 |
0.1091546648 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 11 |
Hb_001534_060 |
0.1139544862 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 12 |
Hb_000049_100 |
0.1162356256 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 13 |
Hb_027380_080 |
0.1168322871 |
- |
- |
hypothetical protein JCGZ_04649 [Jatropha curcas] |
| 14 |
Hb_000008_160 |
0.1181461963 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 15 |
Hb_000948_240 |
0.1182404502 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011282_040 |
0.1202693195 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 17 |
Hb_000185_010 |
0.1215294676 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 18 |
Hb_000808_130 |
0.1216065184 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |
| 19 |
Hb_006198_030 |
0.1220836652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000676_320 |
0.1227479819 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |