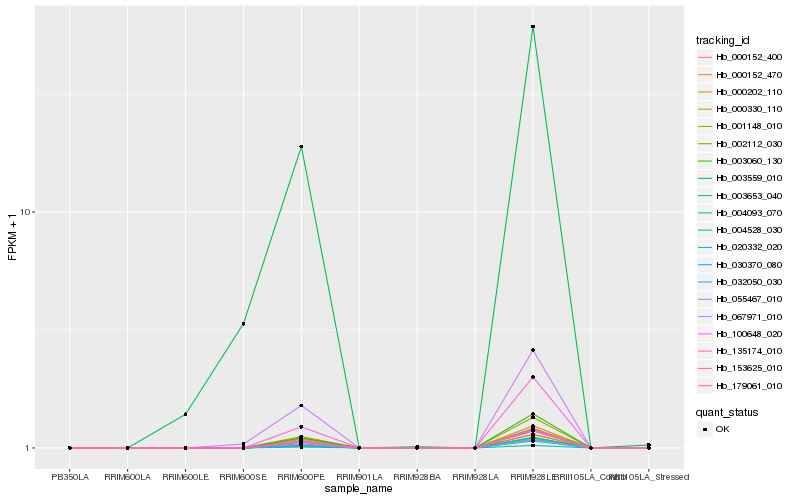
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_067971_010 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_003559_010 |
0.0621564859 |
- |
- |
Peroxidase 27 precursor, putative [Ricinus communis] |
| 3 |
Hb_032050_030 |
0.095324856 |
- |
- |
C, putative [Ricinus communis] |
| 4 |
Hb_030370_080 |
0.0953258931 |
- |
- |
hypothetical protein SELMODRAFT_100988 [Selaginella moellendorffii] |
| 5 |
Hb_002112_030 |
0.0953998238 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_153625_010 |
0.0961520231 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 7 |
Hb_000152_400 |
0.096554775 |
- |
- |
PREDICTED: uncharacterized protein LOC105648582 [Jatropha curcas] |
| 8 |
Hb_020332_020 |
0.0969786795 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 9 |
Hb_100648_020 |
0.0973303515 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 10 |
Hb_004093_070 |
0.0977940137 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 11 |
Hb_179061_010 |
0.1000301816 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 12 |
Hb_000202_110 |
0.1012352694 |
- |
- |
PREDICTED: uncharacterized protein LOC103948916 [Pyrus x bretschneideri] |
| 13 |
Hb_003060_130 |
0.1014015307 |
- |
- |
PREDICTED: protein P21-like [Jatropha curcas] |
| 14 |
Hb_000330_110 |
0.1045257496 |
- |
- |
- |
| 15 |
Hb_000152_470 |
0.1051596172 |
- |
- |
- |
| 16 |
Hb_001148_010 |
0.1078744359 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_055467_010 |
0.1093087756 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 18 |
Hb_003653_040 |
0.1093931531 |
- |
- |
hypothetical protein SORBIDRAFT_05g024215 [Sorghum bicolor] |
| 19 |
Hb_135174_010 |
0.1117194594 |
- |
- |
Glycosyl hydrolase family protein 43 isoform 2 [Theobroma cacao] |
| 20 |
Hb_004528_030 |
0.1117323373 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |