| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
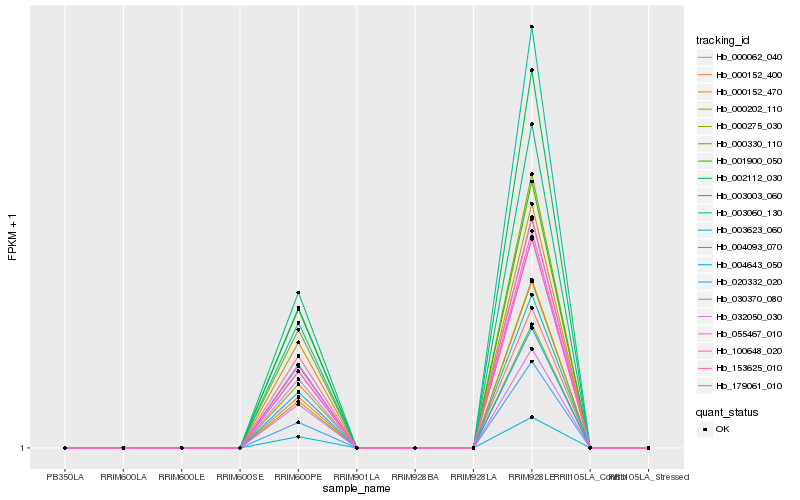
Hb_179061_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 2 |
Hb_004093_070 |
0.0081938037 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 3 |
Hb_100648_020 |
0.010257404 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 4 |
Hb_000330_110 |
0.0123738901 |
- |
- |
- |
| 5 |
Hb_000152_470 |
0.0138837458 |
- |
- |
- |
| 6 |
Hb_153625_010 |
0.0166921799 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 7 |
Hb_055467_010 |
0.0229232169 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 8 |
Hb_002112_030 |
0.0230119371 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_030370_080 |
0.0239132583 |
- |
- |
hypothetical protein SELMODRAFT_100988 [Selaginella moellendorffii] |
| 10 |
Hb_003003_060 |
0.0316877059 |
- |
- |
- |
| 11 |
Hb_004643_050 |
0.0340763001 |
- |
- |
hypothetical protein B456_001G165500 [Gossypium raimondii] |
| 12 |
Hb_032050_030 |
0.0387734074 |
- |
- |
C, putative [Ricinus communis] |
| 13 |
Hb_000152_400 |
0.0484619933 |
- |
- |
PREDICTED: uncharacterized protein LOC105648582 [Jatropha curcas] |
| 14 |
Hb_020332_020 |
0.0507258847 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 15 |
Hb_001900_050 |
0.0511931902 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 16 |
Hb_000275_030 |
0.0547963174 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003623_060 |
0.0648416026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000062_040 |
0.065752855 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 19 |
Hb_000202_110 |
0.0663409512 |
- |
- |
PREDICTED: uncharacterized protein LOC103948916 [Pyrus x bretschneideri] |
| 20 |
Hb_003060_130 |
0.0668217947 |
- |
- |
PREDICTED: protein P21-like [Jatropha curcas] |