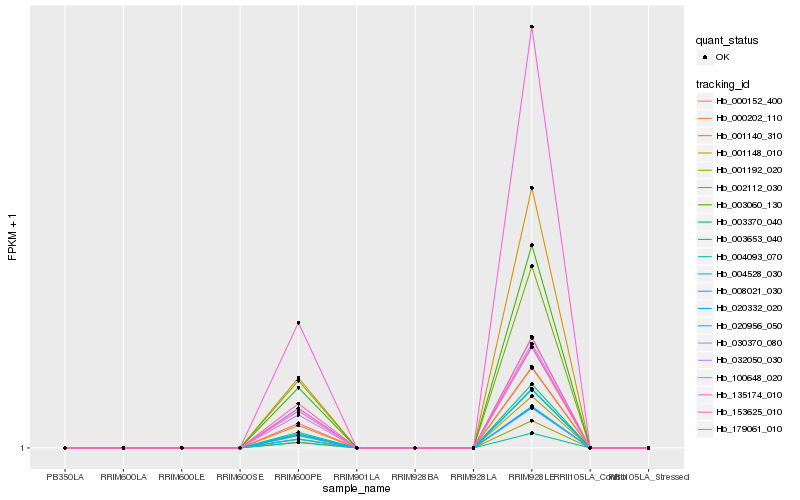
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_400 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648582 [Jatropha curcas] |
| 2 |
Hb_020332_020 |
0.0022672321 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 3 |
Hb_032050_030 |
0.0096993177 |
- |
- |
C, putative [Ricinus communis] |
| 4 |
Hb_000202_110 |
0.0179143014 |
- |
- |
PREDICTED: uncharacterized protein LOC103948916 [Pyrus x bretschneideri] |
| 5 |
Hb_003060_130 |
0.0183963858 |
- |
- |
PREDICTED: protein P21-like [Jatropha curcas] |
| 6 |
Hb_030370_080 |
0.024565138 |
- |
- |
hypothetical protein SELMODRAFT_100988 [Selaginella moellendorffii] |
| 7 |
Hb_002112_030 |
0.0254663988 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_153625_010 |
0.0317844747 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 9 |
Hb_001148_010 |
0.0341567266 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 10 |
Hb_003653_040 |
0.0373097311 |
- |
- |
hypothetical protein SORBIDRAFT_05g024215 [Sorghum bicolor] |
| 11 |
Hb_100648_020 |
0.0382153223 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 12 |
Hb_004093_070 |
0.0402771994 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 13 |
Hb_135174_010 |
0.0418903171 |
- |
- |
Glycosyl hydrolase family protein 43 isoform 2 [Theobroma cacao] |
| 14 |
Hb_004528_030 |
0.0419149444 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 15 |
Hb_001140_310 |
0.0419192013 |
- |
- |
hypothetical protein AMTR_s00025p00025050 [Amborella trichopoda] |
| 16 |
Hb_008021_030 |
0.0428775689 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_020956_050 |
0.045873613 |
- |
- |
PREDICTED: uncharacterized protein LOC102670328 [Glycine max] |
| 18 |
Hb_179061_010 |
0.0484619933 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 19 |
Hb_001192_020 |
0.0570094193 |
- |
- |
- |
| 20 |
Hb_003370_040 |
0.0587139266 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |