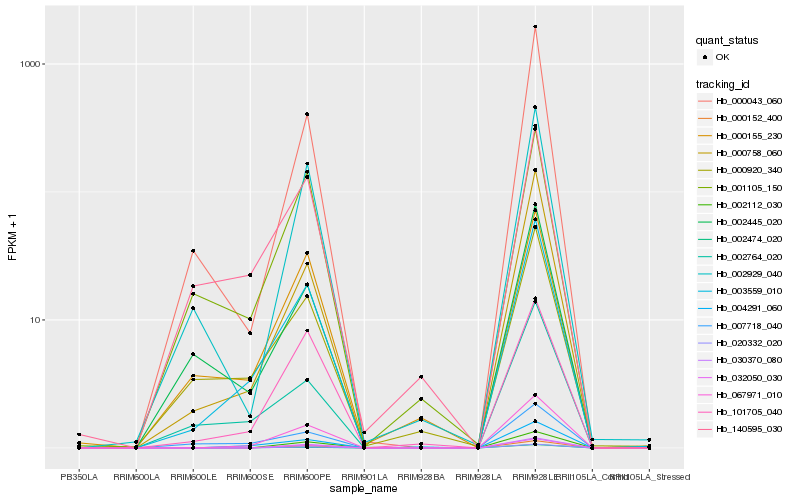
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003559_010 |
0.0 |
- |
- |
Peroxidase 27 precursor, putative [Ricinus communis] |
| 2 |
Hb_067971_010 |
0.0621564859 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 3 |
Hb_000758_060 |
0.1022820866 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45960 [Jatropha curcas] |
| 4 |
Hb_000920_340 |
0.1110131802 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_002474_020 |
0.1157333288 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104585652 [Nelumbo nucifera] |
| 6 |
Hb_004291_060 |
0.1166583288 |
- |
- |
hypothetical protein F775_42457 [Aegilops tauschii] |
| 7 |
Hb_002764_020 |
0.1190493359 |
- |
- |
hypothetical protein JCGZ_24084 [Jatropha curcas] |
| 8 |
Hb_101705_040 |
0.1208485868 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_000043_060 |
0.1222376731 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 10 |
Hb_000155_230 |
0.1240050325 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 11 |
Hb_007718_040 |
0.1264461669 |
- |
- |
hypothetical protein PRUPE_ppb025054mg [Prunus persica] |
| 12 |
Hb_140595_030 |
0.1266354242 |
- |
- |
- |
| 13 |
Hb_002929_040 |
0.1275340763 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_002445_020 |
0.1282944613 |
- |
- |
- |
| 15 |
Hb_001105_150 |
0.1289403138 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 16 |
Hb_000152_400 |
0.1323322121 |
- |
- |
PREDICTED: uncharacterized protein LOC105648582 [Jatropha curcas] |
| 17 |
Hb_032050_030 |
0.1323996874 |
- |
- |
C, putative [Ricinus communis] |
| 18 |
Hb_020332_020 |
0.1324171205 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 19 |
Hb_030370_080 |
0.1338498923 |
- |
- |
hypothetical protein SELMODRAFT_100988 [Selaginella moellendorffii] |
| 20 |
Hb_002112_030 |
0.1339894059 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |