| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
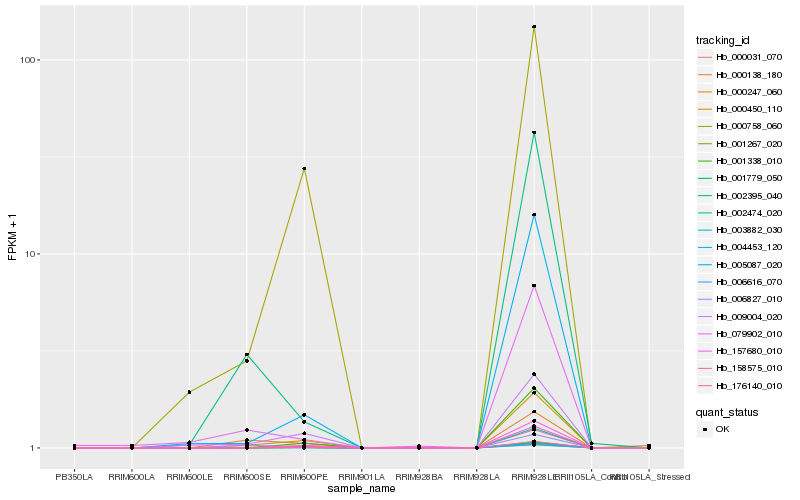
Hb_000450_110 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 2 |
Hb_000247_060 |
0.0443378651 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ORG2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_006616_070 |
0.1177125243 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_001267_020 |
0.1190220181 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 5 |
Hb_002474_020 |
0.1258921989 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104585652 [Nelumbo nucifera] |
| 6 |
Hb_009004_020 |
0.1282620268 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 7 |
Hb_002395_040 |
0.1440930678 |
- |
- |
unknown [Populus trichocarpa] |
| 8 |
Hb_000138_180 |
0.1456615846 |
- |
- |
PREDICTED: uncharacterized protein LOC105131773 [Populus euphratica] |
| 9 |
Hb_005087_020 |
0.1525826364 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_158575_010 |
0.1526942073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_157680_010 |
0.1527289693 |
- |
- |
PREDICTED: uncharacterized protein LOC103444007 [Malus domestica] |
| 12 |
Hb_004453_120 |
0.152927176 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 13 |
Hb_176140_010 |
0.1532357731 |
- |
- |
polyprotein [Oryza australiensis] |
| 14 |
Hb_006827_010 |
0.1532783039 |
- |
- |
retrotransposon protein, putative, unclassified, expressed [Oryza sativa Japonica Group] |
| 15 |
Hb_001779_050 |
0.1533309777 |
- |
- |
polyprotein [Ananas comosus] |
| 16 |
Hb_000758_060 |
0.1537994154 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45960 [Jatropha curcas] |
| 17 |
Hb_001338_010 |
0.1551504292 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 18 |
Hb_003882_030 |
0.1553614554 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 19 |
Hb_079902_010 |
0.1559928921 |
- |
- |
cytochrome c biogenesis B (mitochondrion) [Hevea brasiliensis] |
| 20 |
Hb_000031_070 |
0.1560935242 |
- |
- |
Thioredoxin domain-containing protein, putative [Ricinus communis] |