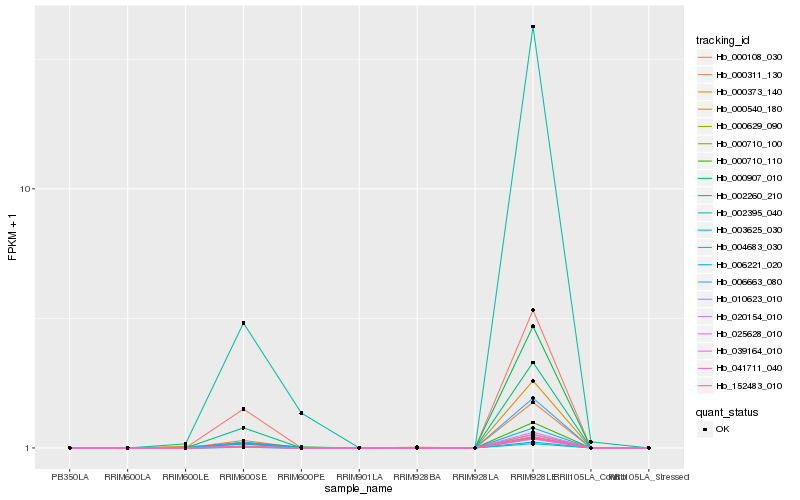
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025628_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104233990 [Nicotiana sylvestris] |
| 2 |
Hb_000540_180 |
0.0004133987 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 3 |
Hb_006663_080 |
0.0280200075 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 4 |
Hb_020154_010 |
0.032243782 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_003625_030 |
0.0462362569 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy subclass [Oryza sativa Japonica Group] |
| 6 |
Hb_004683_030 |
0.04697298 |
- |
- |
PREDICTED: uncharacterized protein LOC105778947 [Gossypium raimondii] |
| 7 |
Hb_000629_090 |
0.0486165232 |
- |
- |
hypothetical protein JCGZ_09480 [Jatropha curcas] |
| 8 |
Hb_039164_010 |
0.0489374843 |
- |
- |
- |
| 9 |
Hb_041711_040 |
0.0492379876 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 10 |
Hb_152483_010 |
0.0506012991 |
- |
- |
hypothetical protein JCGZ_22450 [Jatropha curcas] |
| 11 |
Hb_000710_100 |
0.0736831133 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 12 |
Hb_010623_010 |
0.0748756788 |
- |
- |
PREDICTED: uncharacterized protein LOC105643732 [Jatropha curcas] |
| 13 |
Hb_000311_130 |
0.0785639659 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 14 |
Hb_002395_040 |
0.0813536815 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_000907_010 |
0.0990772584 |
- |
- |
Ankyrin repeat-containing-like protein [Theobroma cacao] |
| 16 |
Hb_002260_210 |
0.1100686264 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000108_030 |
0.1100966756 |
- |
- |
- |
| 18 |
Hb_000710_110 |
0.1185933504 |
- |
- |
Vps51/Vps67 [Citrus endogenous pararetrovirus] |
| 19 |
Hb_006221_020 |
0.1240150222 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 20 |
Hb_000373_140 |
0.1319389569 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |