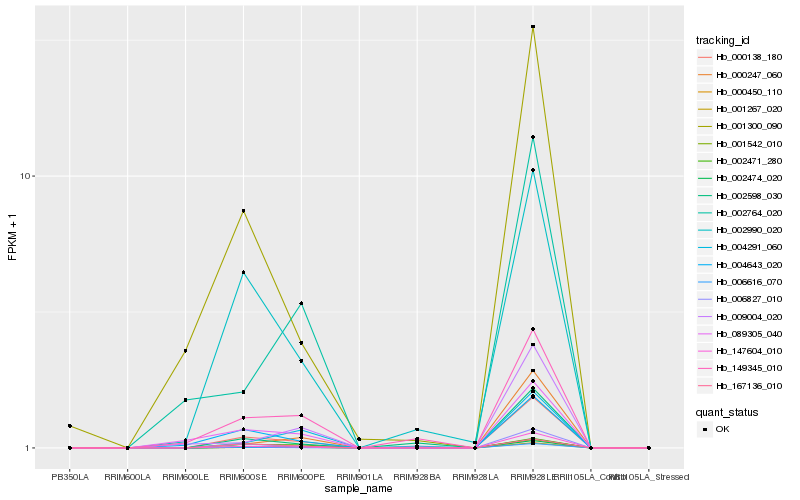
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001267_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 2 |
Hb_000138_180 |
0.0391648007 |
- |
- |
PREDICTED: uncharacterized protein LOC105131773 [Populus euphratica] |
| 3 |
Hb_006616_070 |
0.0745308914 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_006827_010 |
0.0777465199 |
- |
- |
retrotransposon protein, putative, unclassified, expressed [Oryza sativa Japonica Group] |
| 5 |
Hb_002474_020 |
0.0961963798 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104585652 [Nelumbo nucifera] |
| 6 |
Hb_000450_110 |
0.1190220181 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 7 |
Hb_000247_060 |
0.1410019415 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ORG2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004643_020 |
0.1697641452 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 9 |
Hb_149345_010 |
0.1705283949 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 10 |
Hb_002990_020 |
0.1747153597 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 11 |
Hb_147604_010 |
0.1751757061 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 12 |
Hb_001300_090 |
0.1766200524 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 13 |
Hb_009004_020 |
0.1782717145 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 14 |
Hb_004291_060 |
0.1794595489 |
- |
- |
hypothetical protein F775_42457 [Aegilops tauschii] |
| 15 |
Hb_002471_280 |
0.1838922911 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 16 |
Hb_002764_020 |
0.1847176627 |
- |
- |
hypothetical protein JCGZ_24084 [Jatropha curcas] |
| 17 |
Hb_002598_030 |
0.1851443465 |
- |
- |
- |
| 18 |
Hb_089305_040 |
0.1867109306 |
- |
- |
- |
| 19 |
Hb_167136_010 |
0.1883609347 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 20 |
Hb_001542_010 |
0.191879569 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |