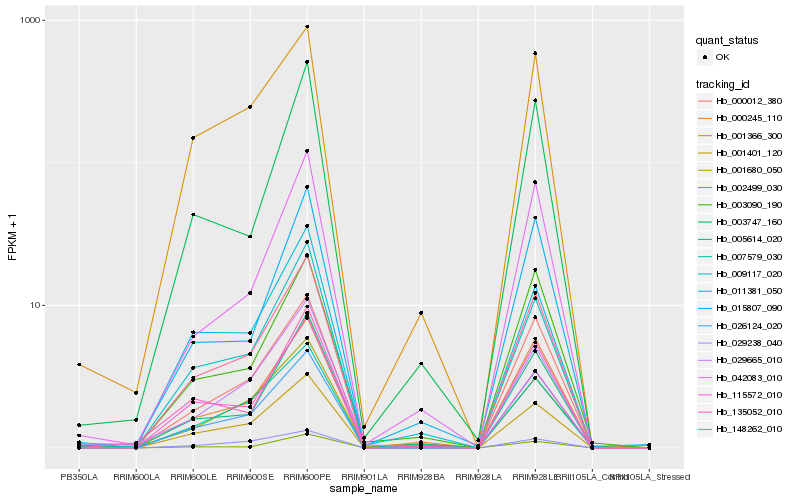
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148262_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_026124_020 |
0.0395207893 |
- |
- |
PREDICTED: uncharacterized protein LOC105644241 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001401_120 |
0.0477446024 |
- |
- |
hypothetical protein JCGZ_04273 [Jatropha curcas] |
| 4 |
Hb_009117_020 |
0.0633345132 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 5 |
Hb_000012_380 |
0.0764804084 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000245_110 |
0.0872727682 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 7 |
Hb_007579_030 |
0.0897886884 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 8 |
Hb_005614_020 |
0.0923410406 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001366_300 |
0.0941904713 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 10 |
Hb_135052_010 |
0.0986357229 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_029665_010 |
0.1028408084 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_011381_050 |
0.104936112 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 13 |
Hb_115572_010 |
0.1051692189 |
- |
- |
PREDICTED: beta-galactosidase 6 [Jatropha curcas] |
| 14 |
Hb_042083_010 |
0.1100588195 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Jatropha curcas] |
| 15 |
Hb_003090_190 |
0.1107885488 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 16 |
Hb_029238_040 |
0.1123529879 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 17 |
Hb_001680_050 |
0.112624191 |
- |
- |
- |
| 18 |
Hb_003747_160 |
0.113262785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002499_030 |
0.1140800941 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 20 |
Hb_015807_090 |
0.1150880228 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |