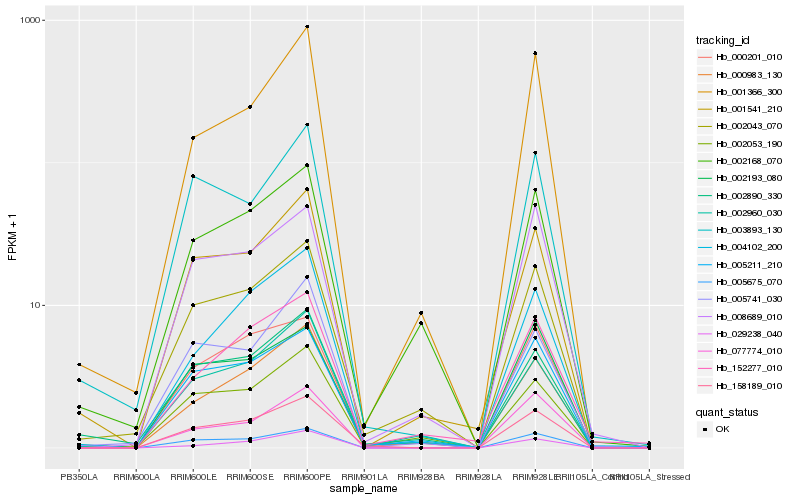
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158189_010 |
0.0 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 2 |
Hb_005675_070 |
0.0834917292 |
- |
- |
hypothetical protein EUGRSUZ_K01585 [Eucalyptus grandis] |
| 3 |
Hb_005211_210 |
0.1000874328 |
- |
- |
lysine and histidine specific transporter family protein [Populus trichocarpa] |
| 4 |
Hb_002043_070 |
0.105707295 |
- |
- |
PREDICTED: jacalin-related lectin 19 [Jatropha curcas] |
| 5 |
Hb_000201_010 |
0.1109286965 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000983_130 |
0.1116103876 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_001541_210 |
0.1194668639 |
- |
- |
hypothetical protein POPTR_0008s19020g [Populus trichocarpa] |
| 8 |
Hb_004102_200 |
0.122953955 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 9 |
Hb_008689_010 |
0.123458057 |
- |
- |
hypothetical protein POPTR_0006s25330g [Populus trichocarpa] |
| 10 |
Hb_077774_010 |
0.1254643617 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 11 |
Hb_002193_080 |
0.1261774696 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 12 |
Hb_002960_030 |
0.1279962479 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 13 |
Hb_001366_300 |
0.1303053676 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 14 |
Hb_003893_130 |
0.1308179751 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_029238_040 |
0.133217106 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 16 |
Hb_002890_330 |
0.1336149597 |
- |
- |
caspase, putative [Ricinus communis] |
| 17 |
Hb_152277_010 |
0.1336892158 |
- |
- |
PREDICTED: 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_002053_190 |
0.1356881484 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005741_030 |
0.1386353411 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 20 |
Hb_002168_070 |
0.1398088782 |
- |
- |
Jasmonate O-methyltransferase, putative [Ricinus communis] |