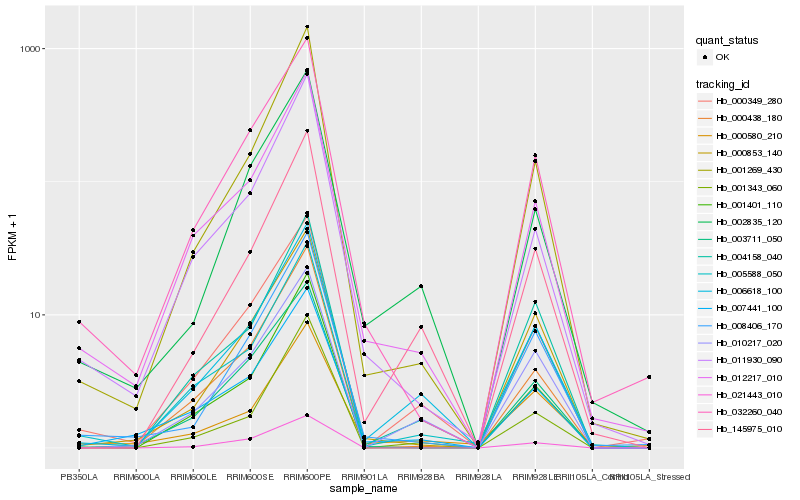
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008406_170 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000853_140 |
0.0684115118 |
- |
- |
hypothetical protein POPTR_0014s12080g [Populus trichocarpa] |
| 3 |
Hb_032260_040 |
0.0832947157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001269_430 |
0.0930181589 |
- |
- |
hypothetical protein JCGZ_04286 [Jatropha curcas] |
| 5 |
Hb_002835_120 |
0.1127152729 |
- |
- |
PREDICTED: uncharacterized protein LOC105640304 [Jatropha curcas] |
| 6 |
Hb_012217_010 |
0.1160715358 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 7 |
Hb_003711_050 |
0.1180953487 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 8 |
Hb_005588_050 |
0.1204102067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004158_040 |
0.1204830572 |
- |
- |
PREDICTED: WAT1-related protein At2g37460-like [Jatropha curcas] |
| 10 |
Hb_000349_280 |
0.120958254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006618_100 |
0.1212841978 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 12 |
Hb_021443_010 |
0.1218506757 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 13 |
Hb_145975_010 |
0.1221990074 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_000580_210 |
0.1245434726 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 15 [Jatropha curcas] |
| 15 |
Hb_007441_100 |
0.1246262975 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 12 [Populus euphratica] |
| 16 |
Hb_000438_180 |
0.1279668277 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_011930_090 |
0.1297091218 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 18 |
Hb_010217_020 |
0.1326004107 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 19 |
Hb_001401_110 |
0.1327761428 |
- |
- |
hypothetical protein JCGZ_04272 [Jatropha curcas] |
| 20 |
Hb_001343_060 |
0.1332363518 |
- |
- |
Cytochrome P450 [Theobroma cacao] |