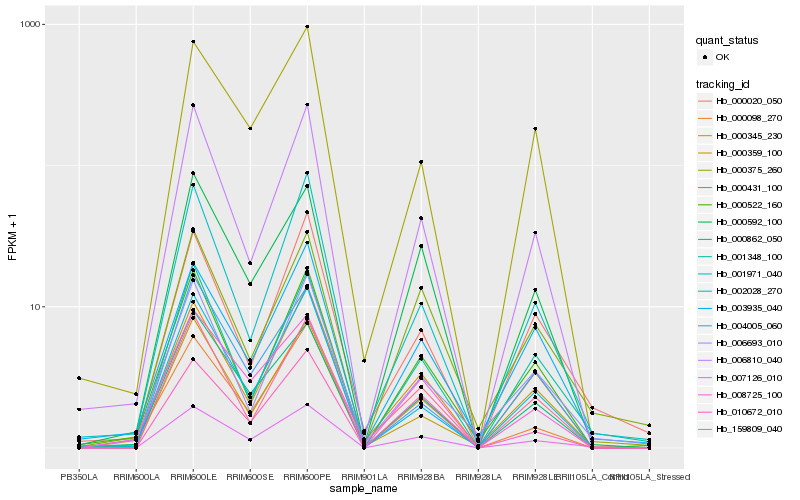
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007126_010 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 2 |
Hb_006810_040 |
0.0985858905 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_001348_100 |
0.1016958201 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 4 |
Hb_000862_050 |
0.103198239 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 5 |
Hb_000592_100 |
0.1036217734 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 6 |
Hb_008725_100 |
0.1107171133 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 7 |
Hb_159809_040 |
0.110883882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000098_270 |
0.1117799947 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 9 |
Hb_001971_040 |
0.1124105049 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 10 |
Hb_010672_010 |
0.1141334558 |
- |
- |
hypothetical protein JCGZ_00895 [Jatropha curcas] |
| 11 |
Hb_002028_270 |
0.1147362688 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000345_230 |
0.1157523196 |
- |
- |
- |
| 13 |
Hb_000375_260 |
0.1178993128 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |
| 14 |
Hb_000522_160 |
0.1195235004 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 15 |
Hb_000020_050 |
0.1210952138 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 16 |
Hb_000359_100 |
0.1234901066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006693_010 |
0.1253015556 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 18 |
Hb_000431_100 |
0.1261233277 |
- |
- |
ribonucleoside-diphosphate reductase small chain, putative [Ricinus communis] |
| 19 |
Hb_004005_060 |
0.1262065409 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 20 |
Hb_003935_040 |
0.1267721775 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |