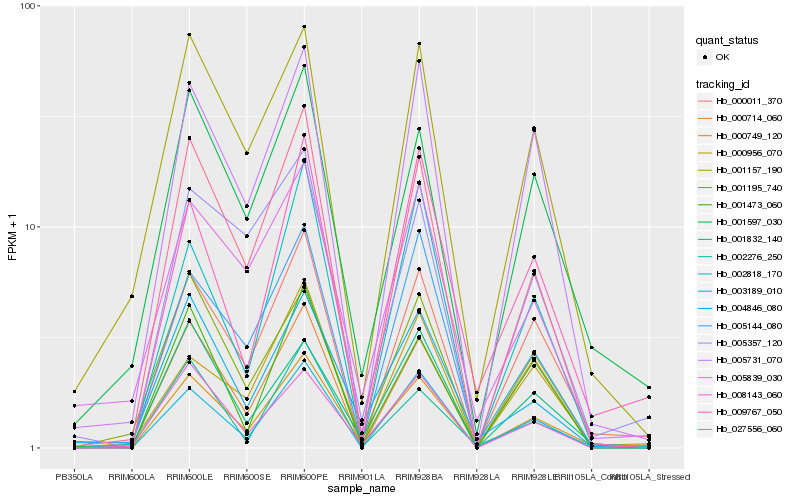
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000749_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644255 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000011_370 |
0.1038064296 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 3 |
Hb_001597_030 |
0.1145167051 |
- |
- |
PREDICTED: uncharacterized protein LOC104445128 isoform X1 [Eucalyptus grandis] |
| 4 |
Hb_027556_060 |
0.1198850784 |
- |
- |
hypothetical protein POPTR_0001s03520g [Populus trichocarpa] |
| 5 |
Hb_008143_060 |
0.1206074802 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 6 |
Hb_005731_070 |
0.1295079533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005144_080 |
0.1337560812 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 8 |
Hb_003189_010 |
0.1359276061 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 9 |
Hb_001195_740 |
0.1388550076 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009767_050 |
0.1414969469 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001157_190 |
0.1445706739 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 12 |
Hb_001473_060 |
0.1446242791 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |
| 13 |
Hb_001832_140 |
0.1447059426 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |
| 14 |
Hb_004846_080 |
0.1452714558 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 15 |
Hb_005357_120 |
0.1454434019 |
transcription factor |
TF Family: MYB-related |
- |
| 16 |
Hb_000714_060 |
0.1479543687 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 1-like [Populus euphratica] |
| 17 |
Hb_002276_250 |
0.1497658235 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002818_170 |
0.1503388031 |
- |
- |
hypothetical protein POPTR_0014s10650g [Populus trichocarpa] |
| 19 |
Hb_005839_030 |
0.1513739786 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |
| 20 |
Hb_000956_070 |
0.152129169 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |