| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
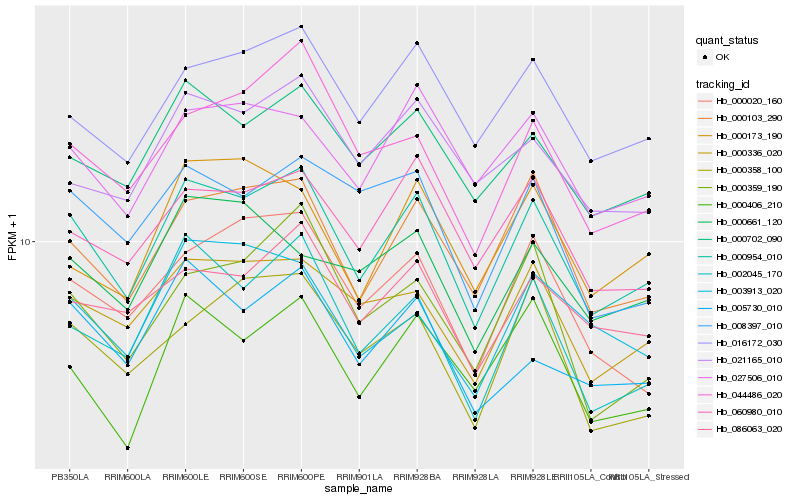
Hb_000954_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 2 |
Hb_000103_290 |
0.0603674909 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 3 |
Hb_000702_090 |
0.0664740732 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 4 |
Hb_016172_030 |
0.0685718193 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 5 |
Hb_000336_020 |
0.0697545315 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 6 |
Hb_002045_170 |
0.0718230961 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 7 |
Hb_060980_010 |
0.0718406598 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 8 |
Hb_086063_020 |
0.0749898894 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 9 |
Hb_027506_010 |
0.0750655671 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 10 |
Hb_044486_020 |
0.0790721152 |
- |
- |
CASTOR protein [Glycine max] |
| 11 |
Hb_000406_210 |
0.0795908 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 12 |
Hb_005730_010 |
0.0800381465 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 13 |
Hb_000020_160 |
0.080059423 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 14 |
Hb_021165_010 |
0.0830440835 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_008397_010 |
0.0830601801 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000359_190 |
0.0848658214 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Nicotiana sylvestris] |
| 17 |
Hb_000173_190 |
0.0861995204 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000358_100 |
0.0862041601 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 19 |
Hb_000661_120 |
0.0871915887 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 20 |
Hb_003913_020 |
0.0871937284 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At5g50170 [Jatropha curcas] |