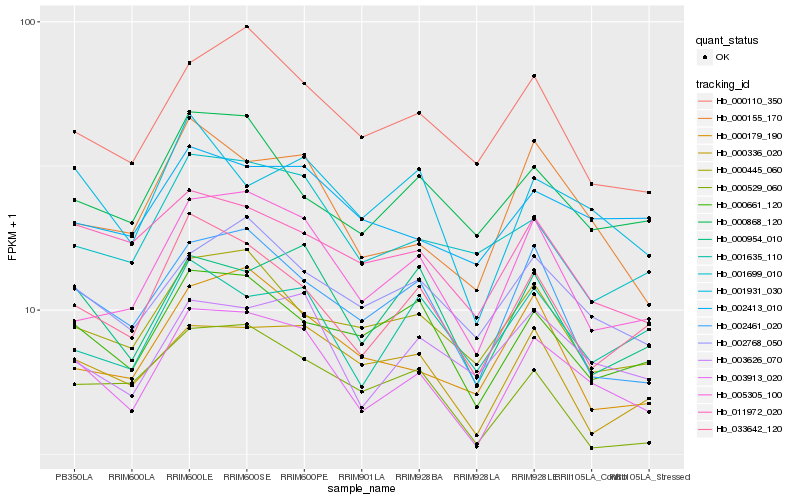
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003913_020 |
0.0 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At5g50170 [Jatropha curcas] |
| 2 |
Hb_002768_050 |
0.0717347962 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 3 |
Hb_003626_070 |
0.0737739208 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 4 |
Hb_033642_120 |
0.0798371537 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 5 |
Hb_002413_010 |
0.0807759428 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_000155_170 |
0.0824829305 |
transcription factor |
TF Family: VOZ |
hypothetical protein EUTSA_v10007498mg [Eutrema salsugineum] |
| 7 |
Hb_000661_120 |
0.0827345905 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 8 |
Hb_000110_350 |
0.0847047773 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 9 |
Hb_000445_060 |
0.0859441084 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000868_120 |
0.0859694542 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 11 |
Hb_005305_100 |
0.0859917609 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000954_010 |
0.0871937284 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 13 |
Hb_001931_030 |
0.0881507877 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta-like [Nelumbo nucifera] |
| 14 |
Hb_002461_020 |
0.0889626848 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 15 |
Hb_011972_020 |
0.0895449708 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_000336_020 |
0.0905345817 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 17 |
Hb_001699_010 |
0.0908923716 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 18 |
Hb_000179_190 |
0.0924306563 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 19 |
Hb_001635_110 |
0.0925053957 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 20 |
Hb_000529_060 |
0.0943285554 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |