| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
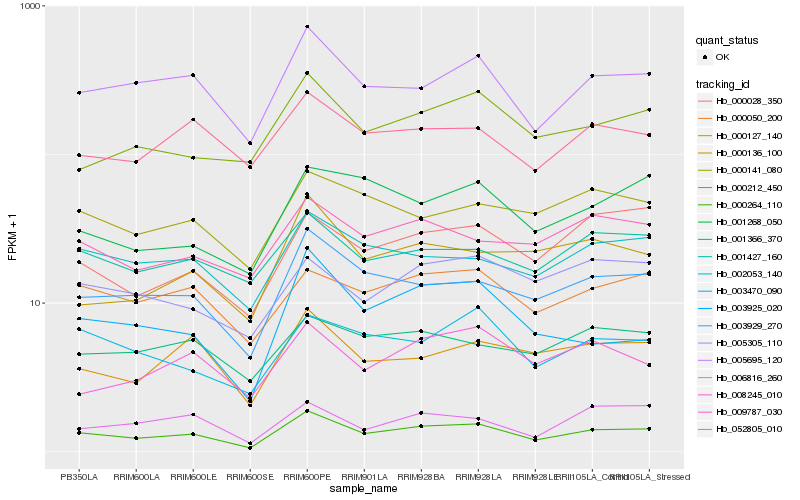
Hb_000264_110 |
0.0 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 13 [Jatropha curcas] |
| 2 |
Hb_003929_270 |
0.0930701142 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_005695_120 |
0.0954833882 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 4 |
Hb_002053_140 |
0.1173773864 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 5 |
Hb_000127_140 |
0.118214031 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 6 |
Hb_000050_200 |
0.1189316619 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 7 |
Hb_003470_090 |
0.124462667 |
- |
- |
PREDICTED: pumilio homolog 12 [Jatropha curcas] |
| 8 |
Hb_000212_450 |
0.126572892 |
- |
- |
PREDICTED: basic leucine zipper and W2 domain-containing protein 2 [Jatropha curcas] |
| 9 |
Hb_001427_160 |
0.1267641965 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_003925_020 |
0.1268575723 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 11 |
Hb_009787_030 |
0.1276321165 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 12 |
Hb_052805_010 |
0.1285226953 |
- |
- |
hypothetical protein POPTR_0009s03650g [Populus trichocarpa] |
| 13 |
Hb_000141_080 |
0.1291328366 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial isoform X1 [Brassica rapa] |
| 14 |
Hb_006816_260 |
0.131337301 |
- |
- |
hypothetical protein JCGZ_23244 [Jatropha curcas] |
| 15 |
Hb_000028_350 |
0.131519612 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |
| 16 |
Hb_001366_370 |
0.1321979363 |
- |
- |
PREDICTED: aarF domain-containing protein kinase 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000136_100 |
0.1325289639 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 18 |
Hb_008245_010 |
0.1331066539 |
- |
- |
PREDICTED: protein NEDD1 [Jatropha curcas] |
| 19 |
Hb_005305_110 |
0.1353272958 |
- |
- |
PREDICTED: uncharacterized protein LOC105648286 [Jatropha curcas] |
| 20 |
Hb_001268_050 |
0.1361796831 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |