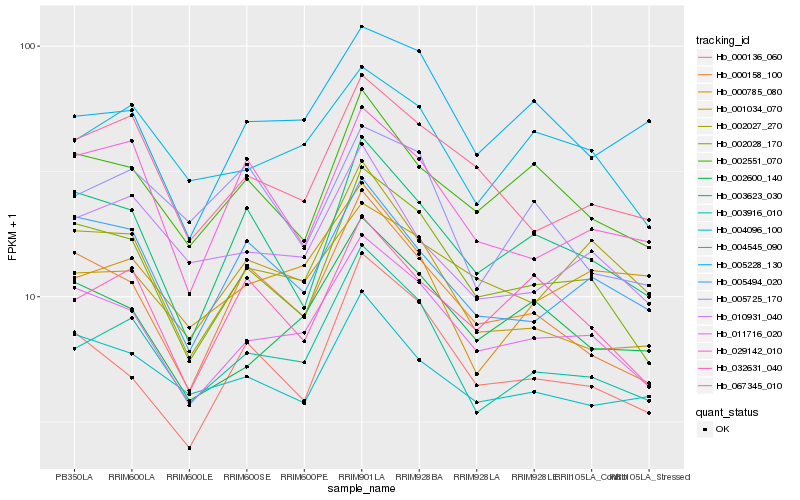
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003916_010 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 2 |
Hb_001034_070 |
0.0870835908 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_011716_020 |
0.090056409 |
- |
- |
PREDICTED: uncharacterized protein LOC105637783 [Jatropha curcas] |
| 4 |
Hb_002028_170 |
0.0984861351 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 5 |
Hb_004545_090 |
0.0998421819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010931_040 |
0.1001060418 |
- |
- |
PREDICTED: nijmegen breakage syndrome 1 protein [Jatropha curcas] |
| 7 |
Hb_000785_080 |
0.1044519139 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 8 |
Hb_067345_010 |
0.1066335087 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 9 |
Hb_000158_100 |
0.1081007971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000136_060 |
0.1100231324 |
- |
- |
hypothetical protein POPTR_0016s06250g [Populus trichocarpa] |
| 11 |
Hb_004096_100 |
0.1114606602 |
- |
- |
PREDICTED: NADPH-dependent diflavin oxidoreductase 1 [Jatropha curcas] |
| 12 |
Hb_005228_130 |
0.1131183509 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_002600_140 |
0.1134340867 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 14 |
Hb_002551_070 |
0.113506562 |
- |
- |
hypothetical protein POPTR_0019s10130g [Populus trichocarpa] |
| 15 |
Hb_005725_170 |
0.1139429637 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 16 |
Hb_002027_270 |
0.1148035689 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 17 |
Hb_029142_010 |
0.1152688266 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 18 |
Hb_005494_020 |
0.1154224386 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 19 |
Hb_003623_030 |
0.1160543943 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 14 [Jatropha curcas] |
| 20 |
Hb_032631_040 |
0.1173131471 |
- |
- |
PREDICTED: uncharacterized protein LOC105638769 [Jatropha curcas] |