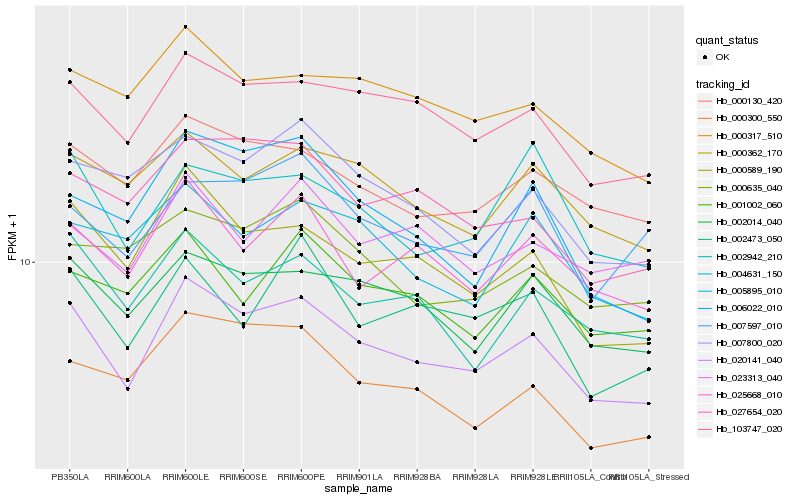
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020141_040 |
0.0 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_103747_020 |
0.0694008216 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 3 |
Hb_000589_190 |
0.0713626821 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002014_040 |
0.0771936825 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 5 |
Hb_000130_420 |
0.082386472 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 6 |
Hb_025668_010 |
0.0824009593 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_002942_210 |
0.0858803866 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 8 |
Hb_027654_020 |
0.0874546436 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 9 |
Hb_000317_510 |
0.0892385676 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 10 |
Hb_000362_170 |
0.0901074087 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 11 |
Hb_007800_020 |
0.0912635144 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006022_010 |
0.0917754057 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 13 |
Hb_000300_550 |
0.0926516481 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 14 |
Hb_005895_010 |
0.0948291376 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002473_050 |
0.0959273864 |
- |
- |
catalytic, putative [Ricinus communis] |
| 16 |
Hb_004631_150 |
0.0979400084 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001002_060 |
0.0981544087 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 18 |
Hb_007597_010 |
0.0986896484 |
- |
- |
hypothetical protein B456_010G072300 [Gossypium raimondii] |
| 19 |
Hb_023313_040 |
0.098770916 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000635_040 |
0.0999874417 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |