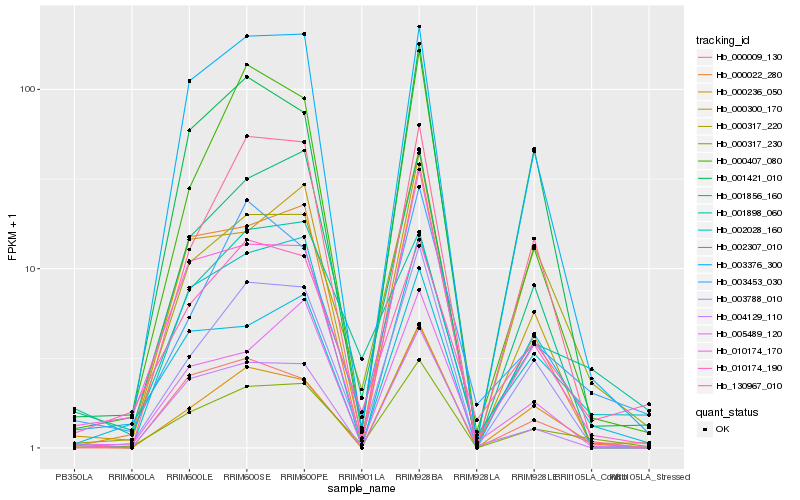
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_230 |
0.0 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 2 |
Hb_003788_010 |
0.1219003469 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 3 |
Hb_000317_220 |
0.129586827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002028_160 |
0.1336587558 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Populus euphratica] |
| 5 |
Hb_001856_160 |
0.1402493072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000022_280 |
0.1419658529 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 7 |
Hb_003376_300 |
0.1440004507 |
- |
- |
Aquaporin PIP2.2, putative [Ricinus communis] |
| 8 |
Hb_010174_190 |
0.14534573 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 9 |
Hb_010174_170 |
0.1458483516 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000236_050 |
0.145849156 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 11 |
Hb_000009_130 |
0.1494445894 |
- |
- |
hypothetical protein POPTR_0007s12410g [Populus trichocarpa] |
| 12 |
Hb_000300_170 |
0.1505552314 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 13 |
Hb_003453_030 |
0.1525166852 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 14 |
Hb_004129_110 |
0.1549329135 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 15 |
Hb_002307_010 |
0.1556748979 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 16 |
Hb_001898_060 |
0.1580866519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005489_120 |
0.1591783833 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 18 |
Hb_001421_010 |
0.1593155966 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000407_080 |
0.1609546406 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 20 |
Hb_130967_010 |
0.1628074504 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |