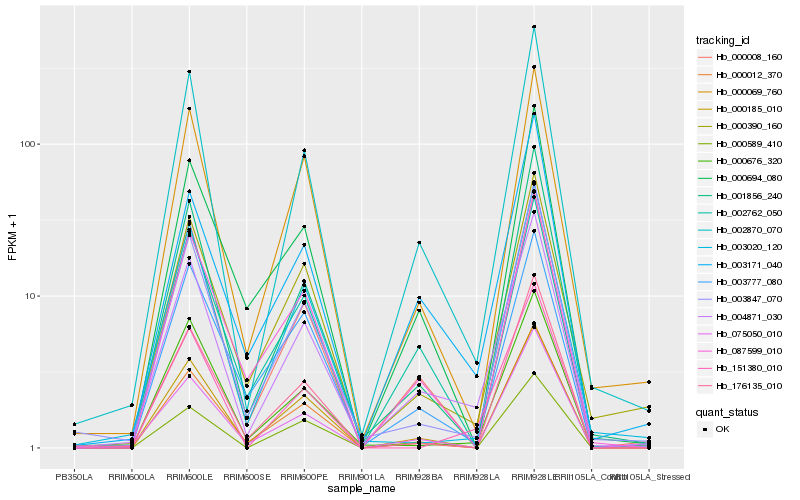
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004871_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002870_070 |
0.0667396747 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 3 |
Hb_003847_070 |
0.1032091307 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001856_240 |
0.1042873424 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 5 |
Hb_000008_160 |
0.1056529177 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 6 |
Hb_000185_010 |
0.1058281515 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 7 |
Hb_003171_040 |
0.1063214245 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 8 |
Hb_000069_760 |
0.1086553079 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000390_160 |
0.1145026426 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 10 |
Hb_000012_370 |
0.1180319084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000676_320 |
0.120410622 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |
| 12 |
Hb_002762_050 |
0.1206675027 |
- |
- |
PREDICTED: uncharacterized protein LOC105640359 [Jatropha curcas] |
| 13 |
Hb_087599_010 |
0.1216345043 |
- |
- |
PREDICTED: tropinone reductase homolog [Solanum tuberosum] |
| 14 |
Hb_075050_010 |
0.1221252914 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000694_080 |
0.1231337267 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 16 |
Hb_003020_120 |
0.1237465141 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 17 |
Hb_000589_410 |
0.1255637224 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic-like isoform X1 [Populus euphratica] |
| 18 |
Hb_151380_010 |
0.1255698446 |
- |
- |
ribosomal protein S4 [Hevea brasiliensis] |
| 19 |
Hb_176135_010 |
0.1256843563 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 20 |
Hb_003777_080 |
0.1261250202 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |