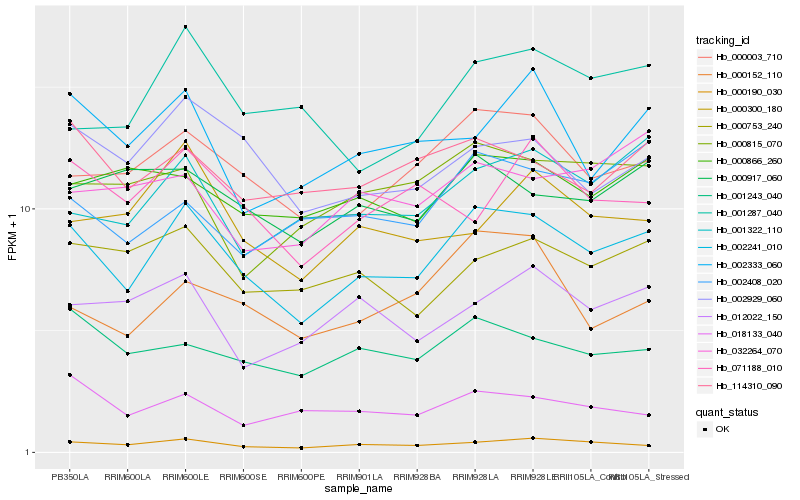
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002241_010 |
0.0 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 2 |
Hb_000003_710 |
0.0949938147 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 3 |
Hb_000190_030 |
0.0953214239 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA-like [Pyrus x bretschneideri] |
| 4 |
Hb_002929_060 |
0.1107779626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001243_040 |
0.1121820932 |
- |
- |
PREDICTED: uncharacterized protein LOC104104316 [Nicotiana tomentosiformis] |
| 6 |
Hb_114310_090 |
0.1133443385 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase [Jatropha curcas] |
| 7 |
Hb_000753_240 |
0.1159011206 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 8 |
Hb_001322_110 |
0.1171188572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000917_060 |
0.1207689614 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 10 |
Hb_000866_260 |
0.121569101 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002333_060 |
0.1233195183 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_001287_040 |
0.1248191275 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 13 |
Hb_000152_110 |
0.1260679482 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_000815_070 |
0.1277399928 |
- |
- |
PREDICTED: methyltransferase-like protein 7A [Jatropha curcas] |
| 15 |
Hb_002408_020 |
0.1277559471 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |
| 16 |
Hb_012022_150 |
0.1280063419 |
- |
- |
PREDICTED: protein root UVB sensitive 5 [Jatropha curcas] |
| 17 |
Hb_018133_040 |
0.1291497521 |
- |
- |
PREDICTED: probable apyrase 7 [Jatropha curcas] |
| 18 |
Hb_000300_180 |
0.1292511305 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 19 |
Hb_071188_010 |
0.1303844767 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 20 |
Hb_032264_070 |
0.1319464282 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |