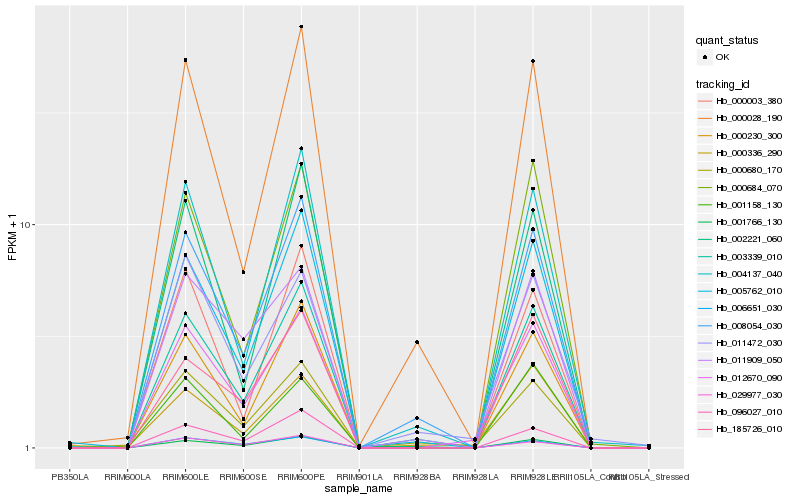
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012670_090 |
0.0 |
- |
- |
GDSL-motif lipase/hydrolase family protein [Populus trichocarpa] |
| 2 |
Hb_001766_130 |
0.0832509339 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_003339_010 |
0.0988189427 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 4 |
Hb_000230_300 |
0.0995907716 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 5 |
Hb_005762_010 |
0.1001991878 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_000684_070 |
0.1085596827 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X1 [Jatropha curcas] |
| 7 |
Hb_008054_030 |
0.108602442 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 8 |
Hb_000680_170 |
0.1141932676 |
- |
- |
hypothetical protein JCGZ_24789 [Jatropha curcas] |
| 9 |
Hb_096027_010 |
0.118270958 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_011472_030 |
0.1200967041 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 11 |
Hb_006651_030 |
0.1207134251 |
- |
- |
unnamed protein product [Homo sapiens] |
| 12 |
Hb_004137_040 |
0.1214764033 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 13 |
Hb_185726_010 |
0.1227602385 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 14 |
Hb_011909_050 |
0.1236916668 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 15 |
Hb_000028_190 |
0.1239811093 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 16 |
Hb_000003_380 |
0.1256384465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002221_060 |
0.1256709408 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 18 |
Hb_000336_290 |
0.1272451856 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 19 |
Hb_001158_130 |
0.1275318703 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 20 |
Hb_029977_030 |
0.1275891 |
transcription factor |
TF Family: bZIP |
conserved hypothetical protein [Ricinus communis] |