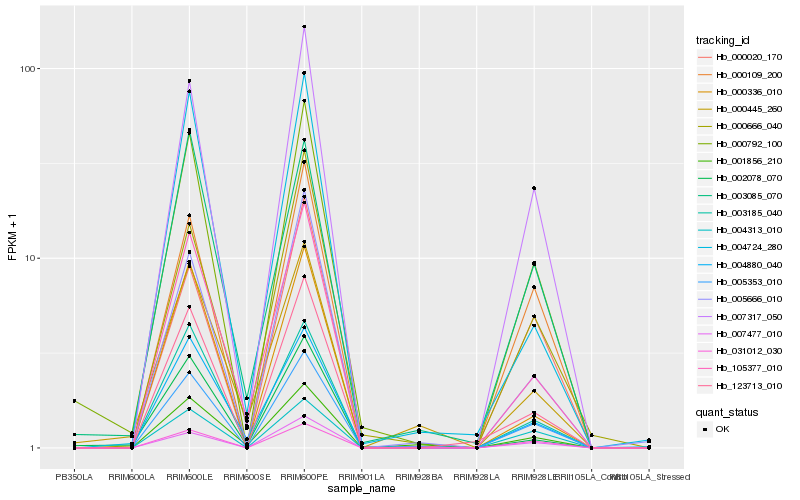
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000792_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940-like [Jatropha curcas] |
| 2 |
Hb_005353_010 |
0.0763737981 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_007317_050 |
0.0947843986 |
- |
- |
hypothetical protein POPTR_0009s16550g [Populus trichocarpa] |
| 4 |
Hb_123713_010 |
0.1049779452 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_031012_030 |
0.1075640491 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 6 |
Hb_000109_200 |
0.1112034236 |
- |
- |
hypothetical protein CISIN_1g037825mg [Citrus sinensis] |
| 7 |
Hb_003185_040 |
0.119078174 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 8 |
Hb_004313_010 |
0.1212809879 |
- |
- |
PREDICTED: patatin-like protein 1 [Prunus mume] |
| 9 |
Hb_105377_010 |
0.1213496509 |
- |
- |
hypothetical protein PRUPE_ppa001414mg [Prunus persica] |
| 10 |
Hb_000336_010 |
0.1279323221 |
transcription factor |
TF Family: C3H |
zinc finger family protein [Populus trichocarpa] |
| 11 |
Hb_002078_070 |
0.1281768565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005666_010 |
0.1291330228 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 13 |
Hb_004880_040 |
0.1321031963 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003085_070 |
0.1337942597 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000666_040 |
0.1357039426 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 16 |
Hb_004724_280 |
0.1381799701 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000020_170 |
0.140391445 |
- |
- |
PREDICTED: aspartic proteinase PCS1 [Jatropha curcas] |
| 18 |
Hb_007477_010 |
0.141514301 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA9 [Phoenix dactylifera] |
| 19 |
Hb_001856_210 |
0.1418504635 |
- |
- |
PREDICTED: uncharacterized protein LOC105644171 [Jatropha curcas] |
| 20 |
Hb_000445_260 |
0.1419135449 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |