| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
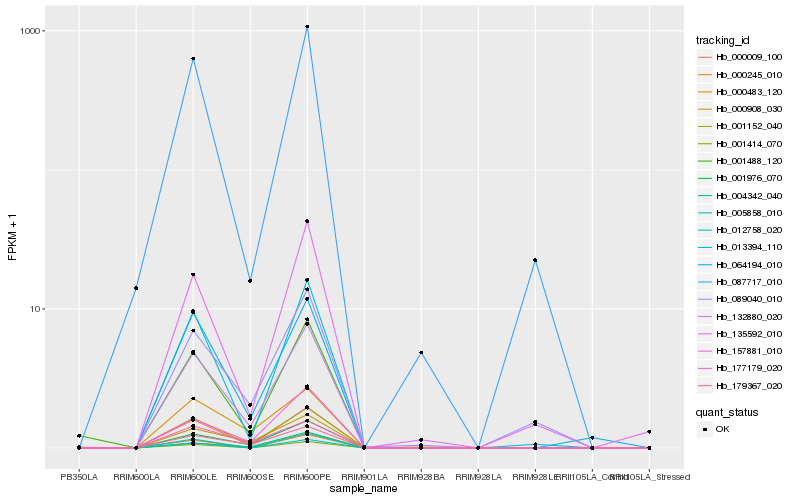
Hb_132880_020 |
0.0 |
- |
- |
hypothetical protein VITISV_026364 [Vitis vinifera] |
| 2 |
Hb_000009_100 |
0.0359551788 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_12260 [Jatropha curcas] |
| 3 |
Hb_001152_040 |
0.0366873526 |
- |
- |
peripheral-type benzodiazepine receptor, putative [Ricinus communis] |
| 4 |
Hb_001976_070 |
0.061829214 |
- |
- |
PREDICTED: uncharacterized protein LOC101497268 [Cicer arietinum] |
| 5 |
Hb_179367_020 |
0.0735400424 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 6 |
Hb_000908_030 |
0.086624843 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 7 |
Hb_157881_010 |
0.0914125495 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_064194_010 |
0.0964608065 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 9 |
Hb_177179_020 |
0.0997439782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_013394_110 |
0.1076038861 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_001488_120 |
0.1077630066 |
- |
- |
hypothetical protein JCGZ_19930 [Jatropha curcas] |
| 12 |
Hb_000483_120 |
0.1231536097 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0004s12530g [Populus trichocarpa] |
| 13 |
Hb_089040_010 |
0.1313105708 |
- |
- |
Glycosyl hydrolase family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_135592_010 |
0.1321981933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004342_040 |
0.1324995512 |
- |
- |
PREDICTED: carotenoid 9,10(9',10')-cleavage dioxygenase 1-like [Jatropha curcas] |
| 16 |
Hb_087717_010 |
0.1356224688 |
- |
- |
hypothetical protein JCGZ_01246 [Jatropha curcas] |
| 17 |
Hb_000245_010 |
0.1391756418 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Populus euphratica] |
| 18 |
Hb_005858_010 |
0.1392028556 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_12260 [Jatropha curcas] |
| 19 |
Hb_012758_020 |
0.1392683799 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 20 |
Hb_001414_070 |
0.139379301 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |