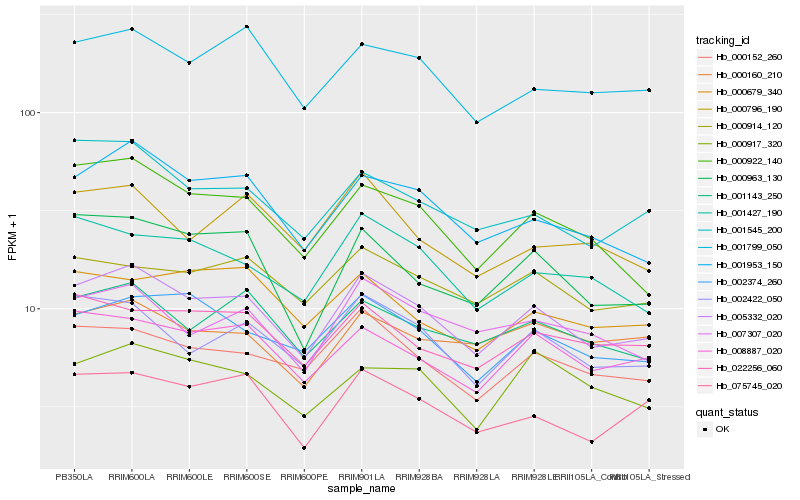
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005332_020 |
0.0 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 2 |
Hb_001953_150 |
0.0665901929 |
- |
- |
PREDICTED: protein DEK isoform X2 [Jatropha curcas] |
| 3 |
Hb_002422_050 |
0.0702371593 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_008887_020 |
0.0707760974 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 5 |
Hb_000152_260 |
0.0717388853 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 6 |
Hb_001143_250 |
0.073610947 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 7 |
Hb_002374_260 |
0.0739788334 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 8 |
Hb_001799_050 |
0.0757614689 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 9 |
Hb_000922_140 |
0.0771536631 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 10 |
Hb_007307_020 |
0.0787490733 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_075745_020 |
0.0807084496 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 12 |
Hb_022256_060 |
0.0807399394 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 13 |
Hb_001545_200 |
0.081141305 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670 [Jatropha curcas] |
| 14 |
Hb_000914_120 |
0.0813042931 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000963_130 |
0.0813708788 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_001427_190 |
0.0837190543 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 17 |
Hb_000917_320 |
0.087976325 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000160_210 |
0.089416298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000796_190 |
0.0914314035 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 20 |
Hb_000679_340 |
0.0920670109 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |