| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
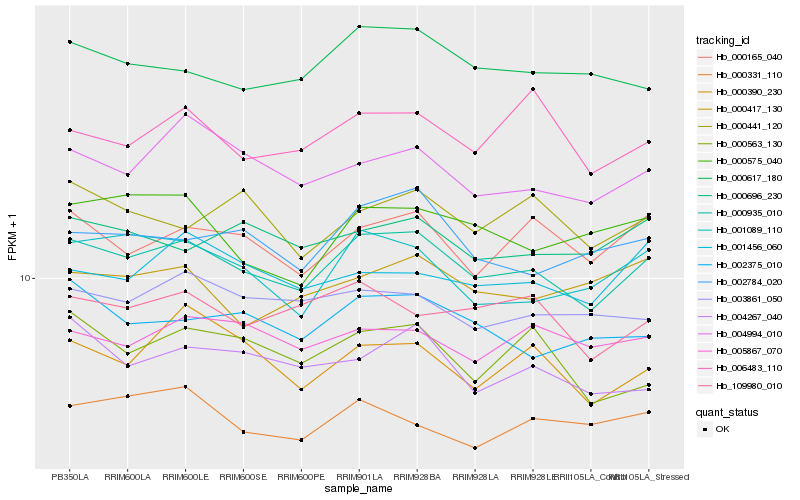
Hb_000935_010 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 2 |
Hb_004994_010 |
0.0625105995 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 3 |
Hb_000165_040 |
0.0630556481 |
- |
- |
hypothetical protein JCGZ_01206 [Jatropha curcas] |
| 4 |
Hb_001089_110 |
0.0637377146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000617_180 |
0.0652053781 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 6 |
Hb_002784_020 |
0.0656988422 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 7 |
Hb_002375_010 |
0.0671803905 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003861_050 |
0.0676938753 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 9 |
Hb_109980_010 |
0.0680882243 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000696_230 |
0.0706226004 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 11 |
Hb_005867_070 |
0.0707253342 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_000575_040 |
0.0727098784 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SC35 [Jatropha curcas] |
| 13 |
Hb_006483_110 |
0.0729760063 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 14 |
Hb_000390_230 |
0.073249633 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 15 |
Hb_004267_040 |
0.0732733148 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 16 |
Hb_001456_060 |
0.073983771 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 17 |
Hb_000441_120 |
0.0741400384 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000563_130 |
0.0744684038 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 19 |
Hb_000417_130 |
0.0750773971 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 20 |
Hb_000331_110 |
0.0759210572 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |