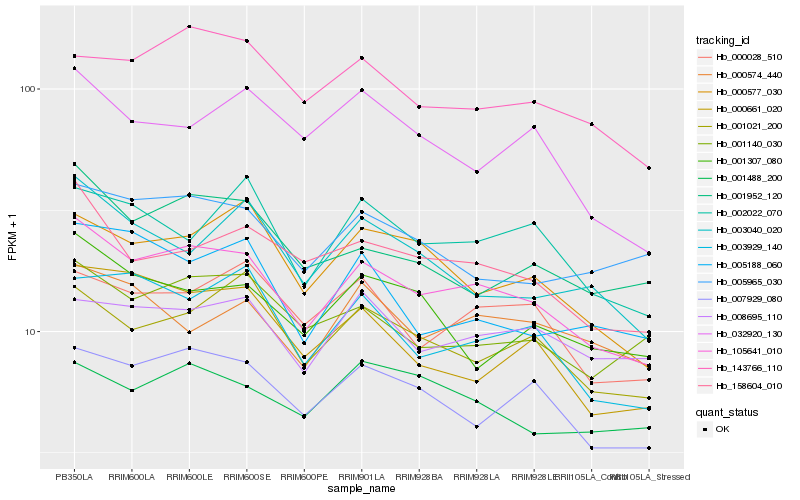
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_105641_010 |
0.0 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 2 |
Hb_007929_080 |
0.0757977656 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 3 |
Hb_158604_010 |
0.0787471196 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 4 |
Hb_001021_200 |
0.0857081397 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 5 |
Hb_003929_140 |
0.0861729073 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000028_510 |
0.0868007311 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 7 |
Hb_143766_110 |
0.0874780494 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 8 |
Hb_000577_030 |
0.0880177236 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 9 |
Hb_001952_120 |
0.089832848 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001140_030 |
0.0906333322 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 11 |
Hb_002022_070 |
0.0916721803 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 12 |
Hb_000661_020 |
0.0950108165 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 13 |
Hb_008695_110 |
0.0956404637 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001307_080 |
0.095956295 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 15 |
Hb_000574_440 |
0.0967097634 |
- |
- |
PREDICTED: uncharacterized protein LOC105634833 [Jatropha curcas] |
| 16 |
Hb_003040_020 |
0.0967658659 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 17 |
Hb_032920_130 |
0.0973486128 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 18 |
Hb_005188_060 |
0.0976515157 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 19 |
Hb_005965_030 |
0.0976633168 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 20 |
Hb_001488_200 |
0.0988578089 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12 homolog isoform X1 [Jatropha curcas] |