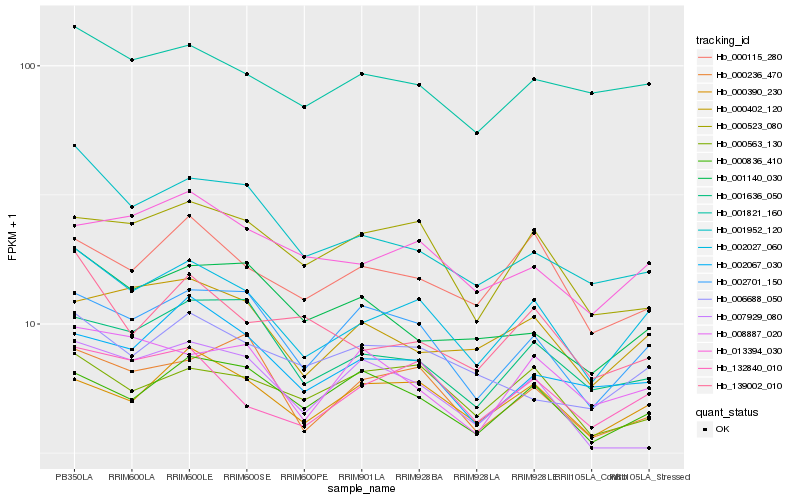
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002027_060 |
0.0 |
- |
- |
PREDICTED: exosome complex component RRP4 [Jatropha curcas] |
| 2 |
Hb_132840_010 |
0.0732291413 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 3 |
Hb_002701_150 |
0.0746384559 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001821_160 |
0.0829190536 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 5 |
Hb_001636_050 |
0.0837582353 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 6 |
Hb_013394_030 |
0.0852569591 |
- |
- |
PREDICTED: exocyst complex component SEC6 [Vitis vinifera] |
| 7 |
Hb_000402_120 |
0.0871044204 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 8 |
Hb_000236_470 |
0.0871504136 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 9 |
Hb_000390_230 |
0.0878793916 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 10 |
Hb_001952_120 |
0.0884992673 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000115_280 |
0.0888651289 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 12 |
Hb_002067_030 |
0.0889257139 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_008887_020 |
0.0941017277 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 14 |
Hb_000523_080 |
0.0954825766 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000836_410 |
0.0961733489 |
- |
- |
sec10, putative [Ricinus communis] |
| 16 |
Hb_001140_030 |
0.0963705509 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 17 |
Hb_139002_010 |
0.0965498981 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000563_130 |
0.0982093613 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 19 |
Hb_007929_080 |
0.0985567779 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 20 |
Hb_006688_050 |
0.0988894434 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX15 isoform X1 [Jatropha curcas] |