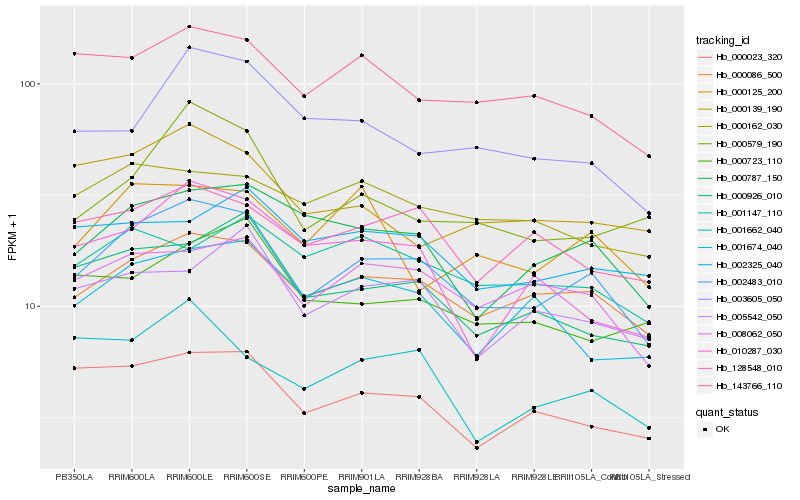
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002483_010 |
0.0 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000086_500 |
0.0683180253 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_008062_050 |
0.090501225 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 4 |
Hb_000787_150 |
0.0960011375 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |
| 5 |
Hb_000023_320 |
0.0962919572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000926_010 |
0.0996879872 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 7 |
Hb_143766_110 |
0.101463832 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 8 |
Hb_128548_010 |
0.107183835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005542_050 |
0.10836417 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 10 |
Hb_001674_040 |
0.1084522641 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 23 [Jatropha curcas] |
| 11 |
Hb_003605_050 |
0.1099040568 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_000579_190 |
0.1100456182 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 13 |
Hb_000139_190 |
0.1141929665 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001662_040 |
0.1143411016 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 15 |
Hb_000162_030 |
0.1153806922 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_010287_030 |
0.1165122891 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000125_200 |
0.1167478886 |
transcription factor |
TF Family: bZIP |
PREDICTED: probable transcription factor PosF21 [Jatropha curcas] |
| 18 |
Hb_002325_040 |
0.1170497165 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001147_110 |
0.1170868149 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 20 |
Hb_000723_110 |
0.1173414261 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |