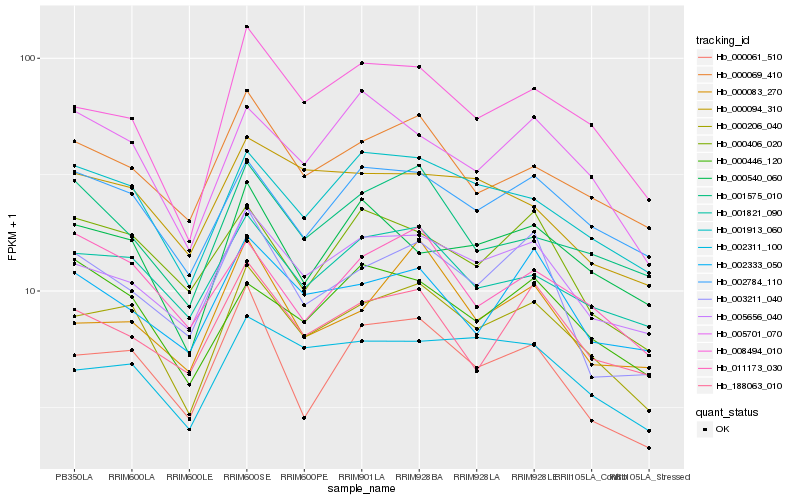
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000206_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 2 |
Hb_008494_010 |
0.0718049166 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 3 |
Hb_005656_040 |
0.0747764717 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 4 |
Hb_001913_060 |
0.0781953472 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 5 |
Hb_001821_090 |
0.0820257104 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 6 |
Hb_002784_110 |
0.0840062952 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 7 |
Hb_002311_100 |
0.0845903808 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 8 |
Hb_000069_410 |
0.0856195156 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 9 |
Hb_000061_510 |
0.0960113042 |
- |
- |
hypothetical protein JCGZ_11686 [Jatropha curcas] |
| 10 |
Hb_005701_070 |
0.0970862381 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 11 |
Hb_188063_010 |
0.0997532231 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 12 |
Hb_000540_060 |
0.1002699542 |
- |
- |
PREDICTED: uncharacterized protein LOC105628177 [Jatropha curcas] |
| 13 |
Hb_000083_270 |
0.101869837 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 14 |
Hb_011173_030 |
0.1021578106 |
- |
- |
pumilio, putative [Ricinus communis] |
| 15 |
Hb_002333_050 |
0.1028534557 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000446_120 |
0.1034526086 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 17 |
Hb_000406_020 |
0.103700687 |
- |
- |
- |
| 18 |
Hb_003211_040 |
0.1046874249 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000094_310 |
0.1047358632 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 20 |
Hb_001575_010 |
0.1048313031 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |