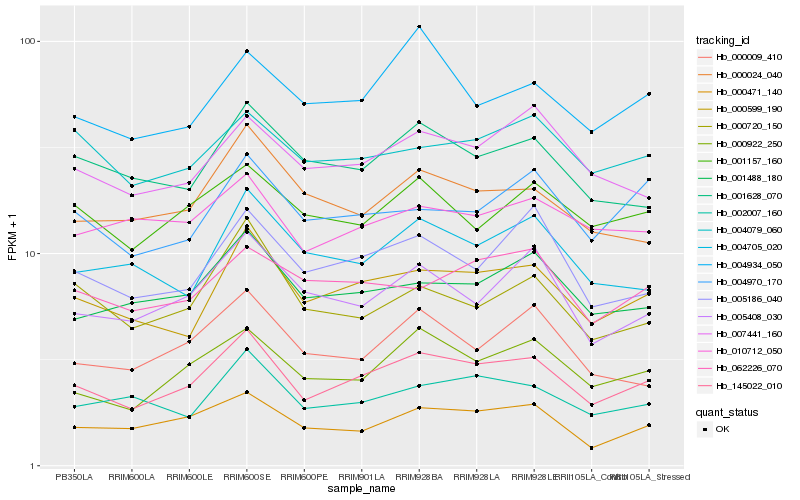
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_145022_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_22699 [Jatropha curcas] |
| 2 |
Hb_000599_190 |
0.0673390753 |
transcription factor |
TF Family: SNF2 |
DNA repair and recombination protein RAD26, putative [Ricinus communis] |
| 3 |
Hb_000922_250 |
0.0785587166 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000471_140 |
0.0872472036 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g28640 [Prunus mume] |
| 5 |
Hb_001488_180 |
0.090643831 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005186_040 |
0.0917519784 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002007_160 |
0.0934365932 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 8 |
Hb_005408_030 |
0.0938244608 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 9 |
Hb_001628_070 |
0.0967005131 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_007441_160 |
0.097158391 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_001157_160 |
0.099972981 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000024_040 |
0.1000050435 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 13 |
Hb_004970_170 |
0.1007204308 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 14 |
Hb_004705_020 |
0.1009314857 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 15 |
Hb_004934_050 |
0.1026654 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 16 |
Hb_062226_070 |
0.1030540722 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000009_410 |
0.1032132906 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 18 |
Hb_004079_060 |
0.1033612941 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 19 |
Hb_000720_150 |
0.1034099438 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X3 [Jatropha curcas] |
| 20 |
Hb_010712_050 |
0.1035424025 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |