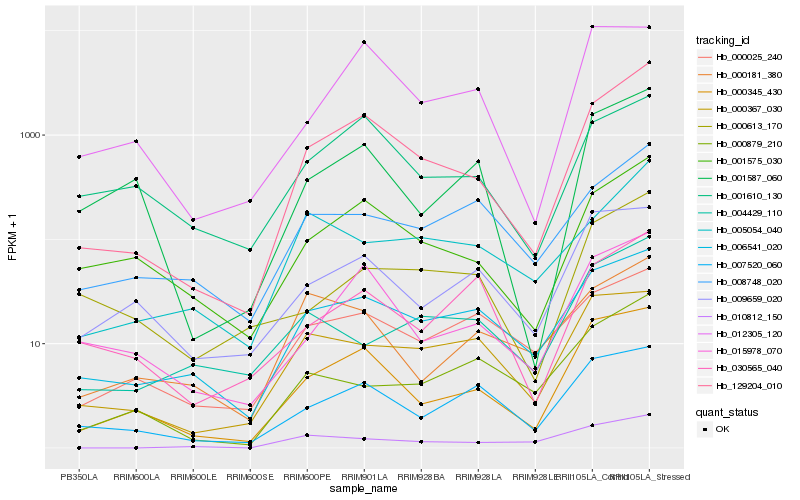
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_129204_010 |
0.0 |
- |
- |
LOC100284182 [Zea mays] |
| 2 |
Hb_000345_430 |
0.1338007111 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 3 |
Hb_001575_030 |
0.155573197 |
- |
- |
hypothetical protein POPTR_0022s00880g [Populus trichocarpa] |
| 4 |
Hb_015978_070 |
0.1594899905 |
- |
- |
- |
| 5 |
Hb_006541_020 |
0.1706906465 |
- |
- |
unknown [Lotus japonicus] |
| 6 |
Hb_001587_060 |
0.1953561905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001610_130 |
0.1978292711 |
- |
- |
40S ribosomal protein S30 [Medicago truncatula] |
| 8 |
Hb_007520_060 |
0.1988551987 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 9 |
Hb_005054_040 |
0.1998439476 |
- |
- |
PREDICTED: 60S ribosomal protein L35a-1 [Gossypium raimondii] |
| 10 |
Hb_010812_150 |
0.2000292763 |
- |
- |
hypothetical protein CICLE_v10009763mg [Citrus clementina] |
| 11 |
Hb_008748_020 |
0.2034393969 |
- |
- |
hypothetical protein B456_005G210700 [Gossypium raimondii] |
| 12 |
Hb_000613_170 |
0.2048697093 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 13 |
Hb_012305_120 |
0.20634885 |
- |
- |
PREDICTED: 60S ribosomal protein L29-1-like [Glycine max] |
| 14 |
Hb_000879_210 |
0.2085102658 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 15 |
Hb_030565_040 |
0.2121694437 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 16 |
Hb_009659_020 |
0.2133981817 |
- |
- |
PREDICTED: uncharacterized protein LOC103870090 [Brassica rapa] |
| 17 |
Hb_000367_030 |
0.2135820288 |
- |
- |
hypothetical protein POPTR_0015s06690g [Populus trichocarpa] |
| 18 |
Hb_000025_240 |
0.2136451115 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 19 |
Hb_004429_110 |
0.2142389718 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 20 |
Hb_000181_380 |
0.2166685661 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |