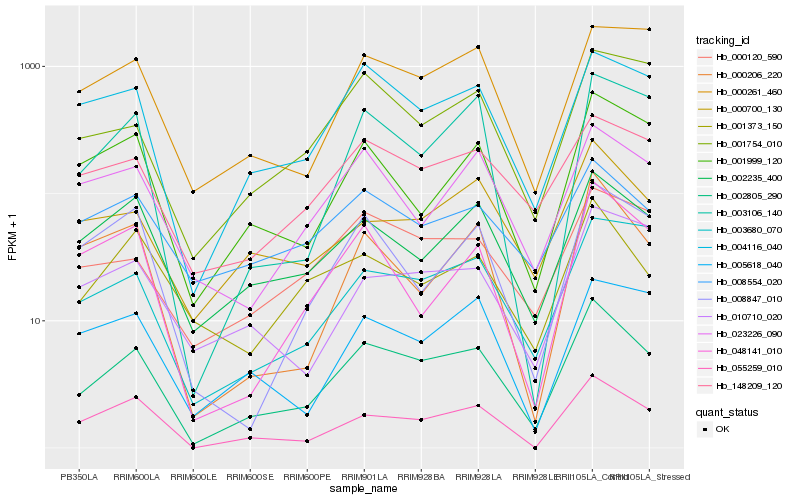
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002805_290 |
0.0 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_055259_010 |
0.130928751 |
- |
- |
- |
| 3 |
Hb_004116_040 |
0.1381373894 |
transcription factor |
TF Family: HMG |
hypothetical protein POPTR_0010s19720g [Populus trichocarpa] |
| 4 |
Hb_002235_400 |
0.1461805636 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 5 |
Hb_003106_140 |
0.1541607898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003680_070 |
0.1547624786 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000700_130 |
0.1550735392 |
- |
- |
PREDICTED: uncharacterized protein At4g13200, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001999_120 |
0.1563953405 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_000120_590 |
0.1572962789 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |
| 10 |
Hb_001754_010 |
0.1587566469 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 11 |
Hb_148209_120 |
0.1592074872 |
- |
- |
hypothetical protein CISIN_1g032846mg [Citrus sinensis] |
| 12 |
Hb_000206_220 |
0.1634229562 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 9 [Jatropha curcas] |
| 13 |
Hb_000261_460 |
0.1661378801 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 14 |
Hb_001373_150 |
0.1666877913 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |
| 15 |
Hb_048141_010 |
0.1671447823 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 2 [Jatropha curcas] |
| 16 |
Hb_023226_090 |
0.1683189978 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 17 |
Hb_008554_020 |
0.1709977468 |
- |
- |
- |
| 18 |
Hb_005618_040 |
0.1732782054 |
- |
- |
- |
| 19 |
Hb_008847_010 |
0.1740060411 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 20 |
Hb_010710_020 |
0.1760076147 |
- |
- |
PREDICTED: uncharacterized protein LOC103322096 [Prunus mume] |