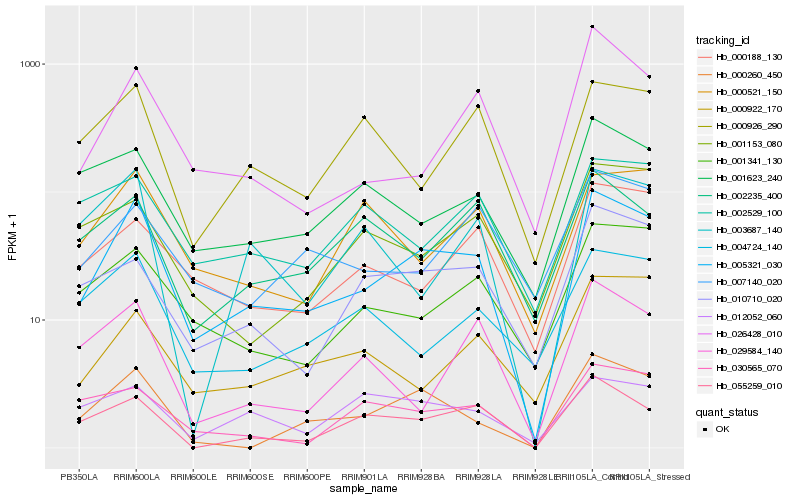
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005321_030 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4 [Jatropha curcas] |
| 2 |
Hb_000260_450 |
0.1671281032 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 3 |
Hb_001341_130 |
0.1858766533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_055259_010 |
0.1864179434 |
- |
- |
- |
| 5 |
Hb_010710_020 |
0.1903273583 |
- |
- |
PREDICTED: uncharacterized protein LOC103322096 [Prunus mume] |
| 6 |
Hb_026428_010 |
0.1908214946 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 7 |
Hb_000188_130 |
0.1946563336 |
- |
- |
PREDICTED: uncharacterized protein LOC102628744 [Citrus sinensis] |
| 8 |
Hb_001623_240 |
0.1955048592 |
- |
- |
hypothetical protein VITISV_004933 [Vitis vinifera] |
| 9 |
Hb_004724_140 |
0.1968623614 |
transcription factor |
TF Family: NF-YB |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 10 |
Hb_000521_150 |
0.199421826 |
- |
- |
PREDICTED: uncharacterized protein LOC105650332 [Jatropha curcas] |
| 11 |
Hb_030565_070 |
0.2003642406 |
- |
- |
- |
| 12 |
Hb_000926_290 |
0.2025593152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007140_020 |
0.2042856031 |
- |
- |
hypothetical protein CICLE_v10008419mg [Citrus clementina] |
| 14 |
Hb_003687_140 |
0.205812811 |
- |
- |
BnaCnng65050D [Brassica napus] |
| 15 |
Hb_012052_060 |
0.2073895522 |
- |
- |
hypothetical protein POPTR_0018s10190g [Populus trichocarpa] |
| 16 |
Hb_002235_400 |
0.2078478469 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000922_170 |
0.2091794911 |
- |
- |
PREDICTED: uncharacterized protein LOC105640383 [Jatropha curcas] |
| 18 |
Hb_029584_140 |
0.2093400254 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 19 |
Hb_002529_100 |
0.2106557479 |
- |
- |
PREDICTED: DET1- and DDB1-associated protein 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001153_080 |
0.2106700616 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL19 [Jatropha curcas] |