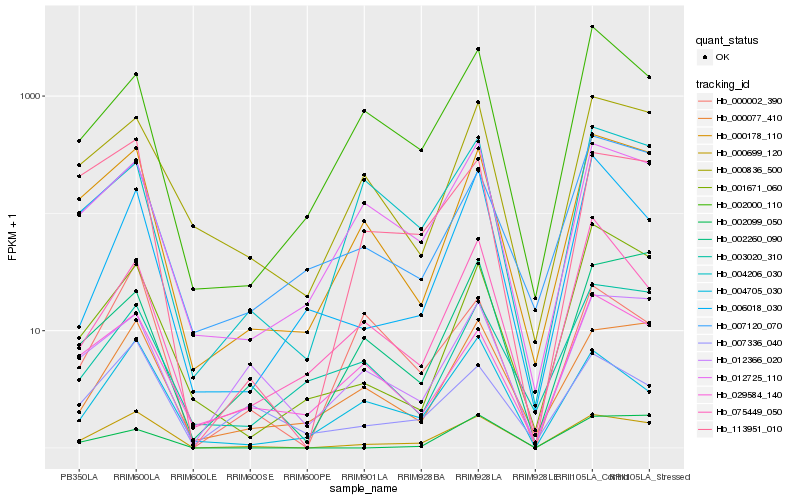
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000699_120 |
0.0 |
- |
- |
DNA-binding family protein [Populus trichocarpa] |
| 2 |
Hb_000178_110 |
0.1479733727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000077_410 |
0.1612997175 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_113951_010 |
0.1628309067 |
- |
- |
hypothetical protein JCGZ_06358 [Jatropha curcas] |
| 5 |
Hb_012725_110 |
0.1762814331 |
- |
- |
hypothetical protein JCGZ_09397 [Jatropha curcas] |
| 6 |
Hb_004705_030 |
0.1782768611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000836_500 |
0.1847954371 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 8 |
Hb_007336_040 |
0.1931423103 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_012366_020 |
0.1939248668 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_007120_070 |
0.1971990612 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 11 |
Hb_002260_090 |
0.1976314632 |
- |
- |
PREDICTED: uncharacterized protein LOC105636025 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001671_060 |
0.200836722 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 13 |
Hb_006018_030 |
0.202088955 |
- |
- |
PREDICTED: uncharacterized protein LOC105108875 isoform X1 [Populus euphratica] |
| 14 |
Hb_004206_030 |
0.2051508118 |
- |
- |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 15 |
Hb_002000_110 |
0.206816733 |
- |
- |
ASR-like protein 2 [Hevea brasiliensis] |
| 16 |
Hb_029584_140 |
0.2091713937 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 17 |
Hb_000002_390 |
0.2091968582 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_075449_050 |
0.2096475589 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 19 |
Hb_002099_050 |
0.211614159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003020_310 |
0.2141741853 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |