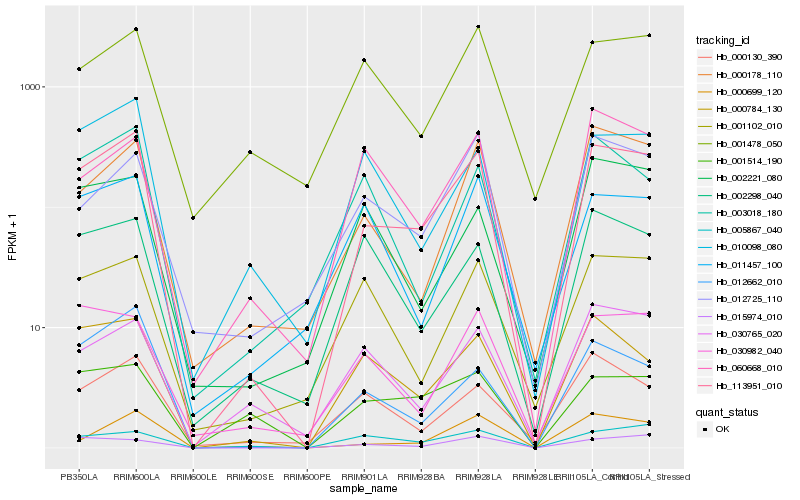
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_113951_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_06358 [Jatropha curcas] |
| 2 |
Hb_000178_110 |
0.1567628581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000784_130 |
0.1581645737 |
- |
- |
hypothetical protein JCGZ_22813 [Jatropha curcas] |
| 4 |
Hb_000699_120 |
0.1628309067 |
- |
- |
DNA-binding family protein [Populus trichocarpa] |
| 5 |
Hb_015974_010 |
0.168918829 |
- |
- |
endonuclease/exonuclease/phosphatase family protein [Medicago truncatula] |
| 6 |
Hb_012662_010 |
0.1700789495 |
- |
- |
hypothetical protein JCGZ_06358 [Jatropha curcas] |
| 7 |
Hb_000130_390 |
0.1707970676 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Pyrus x bretschneideri] |
| 8 |
Hb_010098_080 |
0.1724634054 |
- |
- |
PREDICTED: non-specific phospholipase C4-like [Jatropha curcas] |
| 9 |
Hb_030982_040 |
0.1750983892 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 10 |
Hb_030765_020 |
0.1776958602 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_001102_010 |
0.1790961529 |
- |
- |
PREDICTED: solanesyl diphosphate synthase 3, chloroplastic/mitochondrial-like isoform X2 [Glycine max] |
| 12 |
Hb_012725_110 |
0.1803829826 |
- |
- |
hypothetical protein JCGZ_09397 [Jatropha curcas] |
| 13 |
Hb_005867_040 |
0.1820425542 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein RKD4 [Jatropha curcas] |
| 14 |
Hb_003018_180 |
0.1860666433 |
- |
- |
PREDICTED: WEB family protein At3g51220-like [Jatropha curcas] |
| 15 |
Hb_002298_040 |
0.1870571811 |
- |
- |
PREDICTED: uncharacterized protein LOC105633642 [Jatropha curcas] |
| 16 |
Hb_001514_190 |
0.1884076254 |
- |
- |
- |
| 17 |
Hb_011457_100 |
0.1892611628 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_002221_080 |
0.1920984323 |
- |
- |
PREDICTED: uncharacterized protein LOC105639992 [Jatropha curcas] |
| 19 |
Hb_001478_050 |
0.1928038493 |
- |
- |
PREDICTED: uncharacterized protein LOC105640307 [Jatropha curcas] |
| 20 |
Hb_060668_010 |
0.1946065729 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |