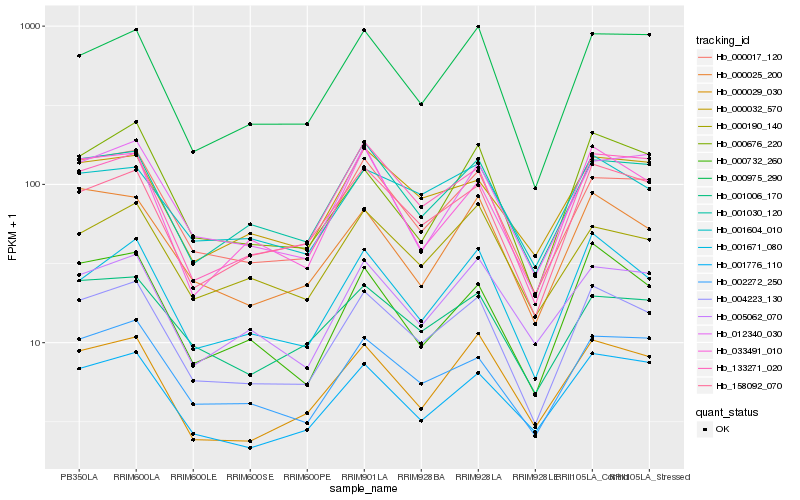
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000017_120 |
0.0647227748 |
- |
- |
PREDICTED: PIN2/TERF1-interacting telomerase inhibitor 1 [Jatropha curcas] |
| 3 |
Hb_158092_070 |
0.0663755999 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_001671_080 |
0.067351766 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_133271_020 |
0.0697094513 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 6 |
Hb_001030_120 |
0.0731594626 |
- |
- |
PREDICTED: tyrosine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 7 |
Hb_002272_250 |
0.0739146274 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000975_290 |
0.0744196722 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 9 |
Hb_001604_010 |
0.0822396297 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |
| 10 |
Hb_000029_030 |
0.0823371721 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 [Jatropha curcas] |
| 11 |
Hb_000732_260 |
0.0838887875 |
- |
- |
PREDICTED: telomerase Cajal body protein 1 [Jatropha curcas] |
| 12 |
Hb_000025_200 |
0.0847479564 |
- |
- |
PREDICTED: cell division control protein 2 homolog A isoform X1 [Jatropha curcas] |
| 13 |
Hb_001776_110 |
0.0851372477 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g07590, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001006_170 |
0.0871615008 |
- |
- |
Endonuclease V, putative [Ricinus communis] |
| 15 |
Hb_000032_570 |
0.0880751227 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: RNA polymerase II transcriptional coactivator KELP [Jatropha curcas] |
| 16 |
Hb_033491_010 |
0.0896315992 |
- |
- |
Phosphatidylinositol N-acetylglucosaminyltransferase subunit C, putative [Ricinus communis] |
| 17 |
Hb_005062_070 |
0.0899872895 |
- |
- |
RNA splicing protein mrs2, mitochondrial, putative [Ricinus communis] |
| 18 |
Hb_000190_140 |
0.0900707762 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 19 |
Hb_012340_030 |
0.0903552598 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 6 [Jatropha curcas] |
| 20 |
Hb_000676_220 |
0.0905137921 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |