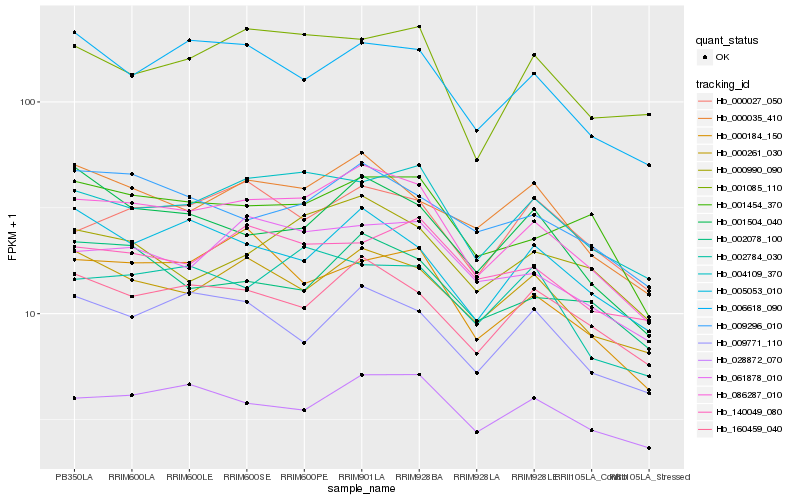
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086287_010 |
0.0 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004109_370 |
0.0736826112 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 3 |
Hb_061878_010 |
0.0780448605 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 4 |
Hb_000261_030 |
0.0788282483 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 5 |
Hb_160459_040 |
0.0802120824 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 6 |
Hb_000035_410 |
0.0811423526 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 7 |
Hb_028872_070 |
0.0824120191 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001454_370 |
0.0833631129 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 9 |
Hb_000990_090 |
0.0861108239 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 10 |
Hb_006618_090 |
0.0864418881 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 11 |
Hb_009296_010 |
0.0894677027 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 12 |
Hb_009771_110 |
0.0922590287 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 13 |
Hb_002784_030 |
0.0926503178 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 14 |
Hb_002078_100 |
0.092919196 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 15 |
Hb_005053_010 |
0.0934612534 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 16 |
Hb_000184_150 |
0.0937947134 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 17 |
Hb_140049_080 |
0.0941876325 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 18 |
Hb_000027_050 |
0.0946673443 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001085_110 |
0.0947657869 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 20 |
Hb_001504_040 |
0.0948144973 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |