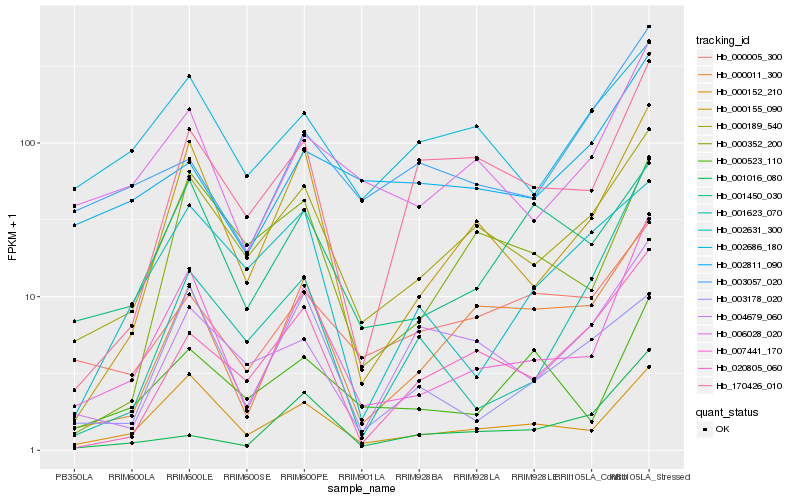
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_170 |
0.0 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 2 |
Hb_000152_210 |
0.1522320255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000155_090 |
0.1527096772 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 4 |
Hb_006028_020 |
0.1777628137 |
- |
- |
PREDICTED: 60S ribosomal protein L38-like [Cicer arietinum] |
| 5 |
Hb_000189_540 |
0.1792811532 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000011_300 |
0.1981212595 |
- |
- |
PREDICTED: proteasome subunit alpha type-5 isoform X1 [Gossypium raimondii] |
| 7 |
Hb_000523_110 |
0.2202982907 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 8 |
Hb_001450_030 |
0.2226039196 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 9 |
Hb_170426_010 |
0.2307099317 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 10 |
Hb_020805_060 |
0.2308916107 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 11 |
Hb_001623_070 |
0.2375887579 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 12 |
Hb_003178_020 |
0.2417490204 |
transcription factor |
TF Family: B3 |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000352_200 |
0.2423433751 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002811_090 |
0.247039423 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 15 |
Hb_002686_180 |
0.2475836064 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 16 |
Hb_000005_300 |
0.247618193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001016_080 |
0.2483480291 |
- |
- |
hypothetical protein EUGRSUZ_J02452 [Eucalyptus grandis] |
| 18 |
Hb_003057_020 |
0.250092408 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |
| 19 |
Hb_004679_060 |
0.2531062048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002631_300 |
0.2545289323 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |