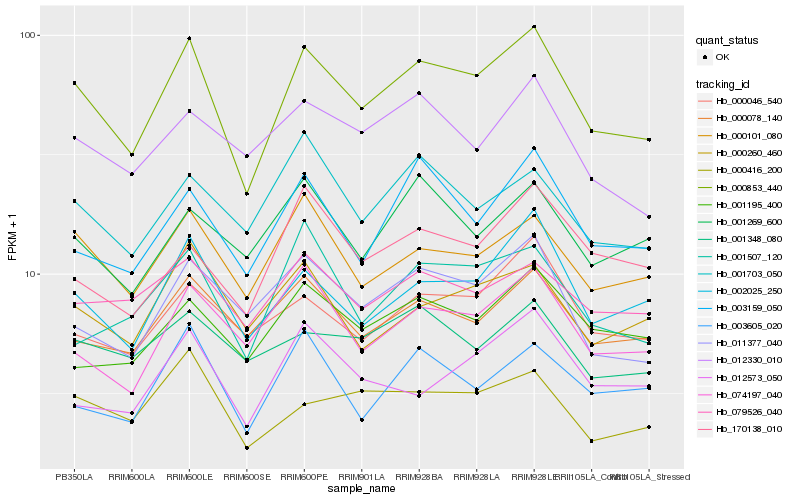
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000853_440 |
0.0 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 2 |
Hb_002025_250 |
0.0847728782 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 3 |
Hb_000101_080 |
0.0880311732 |
- |
- |
PREDICTED: protein CREG1 [Jatropha curcas] |
| 4 |
Hb_000078_140 |
0.0968105965 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 5 |
Hb_011377_040 |
0.0976532321 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 6 |
Hb_003159_050 |
0.0982046269 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_074197_040 |
0.098235316 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 8 |
Hb_000416_200 |
0.1016052096 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 9 |
Hb_000046_540 |
0.1029496854 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 10 |
Hb_003605_020 |
0.1042332417 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 11 |
Hb_000260_460 |
0.1042781588 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 12 |
Hb_079526_040 |
0.1045019297 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 13 |
Hb_001703_050 |
0.1054989808 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 14 |
Hb_001195_400 |
0.1062359632 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 15 |
Hb_012330_010 |
0.1064018574 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001507_120 |
0.1066333162 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_170138_010 |
0.107425311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001269_600 |
0.1085097848 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_001348_080 |
0.1116963664 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 20 |
Hb_012573_050 |
0.1125279826 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |