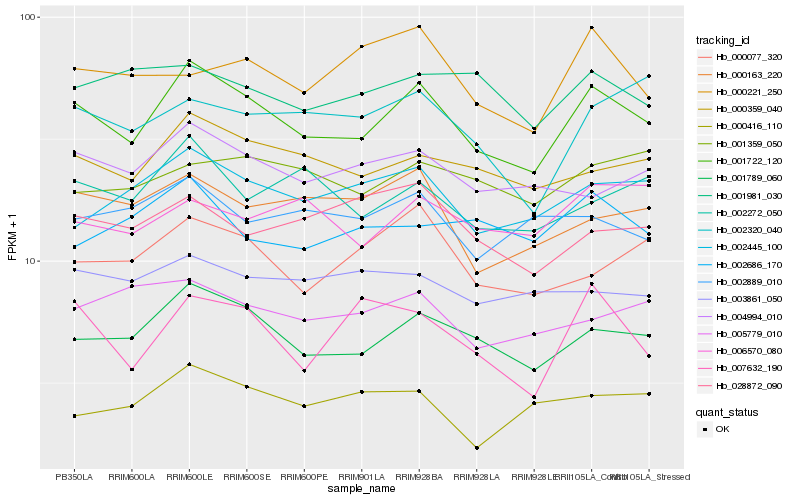
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001722_120 |
0.0 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 2 |
Hb_001789_060 |
0.0628467507 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 3 |
Hb_000359_040 |
0.0849760032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002445_100 |
0.0945088472 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004994_010 |
0.0971009602 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 6 |
Hb_002272_050 |
0.0984268754 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 7 |
Hb_000077_320 |
0.0994238689 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |
| 8 |
Hb_000163_220 |
0.0994839019 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 9 |
Hb_005779_010 |
0.104753976 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_002889_010 |
0.1061280882 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 11 |
Hb_002320_040 |
0.106636956 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |
| 12 |
Hb_001981_030 |
0.106794484 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_007632_190 |
0.1073482253 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic isoform X5 [Camelina sativa] |
| 14 |
Hb_028872_090 |
0.1076589756 |
- |
- |
PREDICTED: urease [Jatropha curcas] |
| 15 |
Hb_001359_050 |
0.1077332166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003861_050 |
0.1080870654 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 17 |
Hb_006570_080 |
0.1085148441 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 18 |
Hb_002686_170 |
0.1093231121 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000416_110 |
0.1096355261 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000221_250 |
0.1101395042 |
- |
- |
hypothetical protein JCGZ_15814 [Jatropha curcas] |