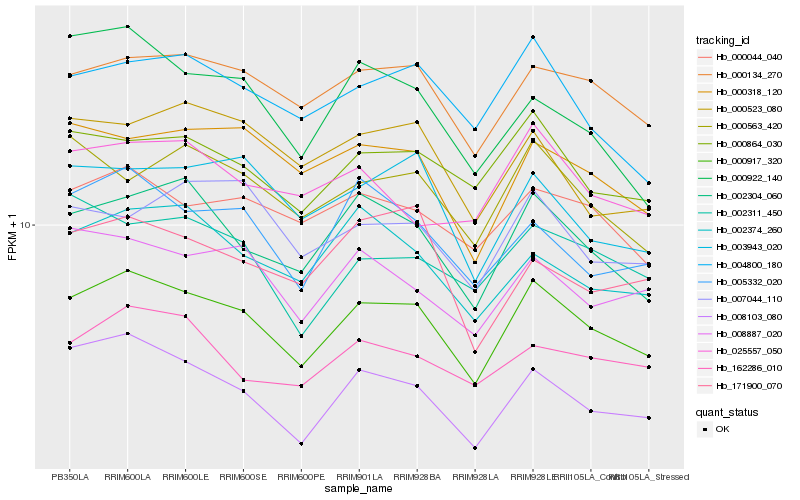
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000917_320 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002304_060 |
0.0711249552 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 3 |
Hb_000134_270 |
0.0753412859 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 4 |
Hb_000864_030 |
0.0822485098 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000044_040 |
0.0862736669 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 6 |
Hb_000318_120 |
0.0862790404 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_004800_180 |
0.0867994036 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 8 |
Hb_005332_020 |
0.087976325 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 9 |
Hb_008887_020 |
0.088818385 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 10 |
Hb_000563_420 |
0.0910569002 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 11 |
Hb_003943_020 |
0.0913984618 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002374_260 |
0.0927070268 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 13 |
Hb_162286_010 |
0.0930984097 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 14 |
Hb_008103_080 |
0.0942804728 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002311_450 |
0.097337076 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 16 |
Hb_000523_080 |
0.097972738 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 17 |
Hb_007044_110 |
0.0987570355 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000922_140 |
0.100697061 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 19 |
Hb_025557_050 |
0.1020923487 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 20 |
Hb_171900_070 |
0.1029332212 |
- |
- |
- |