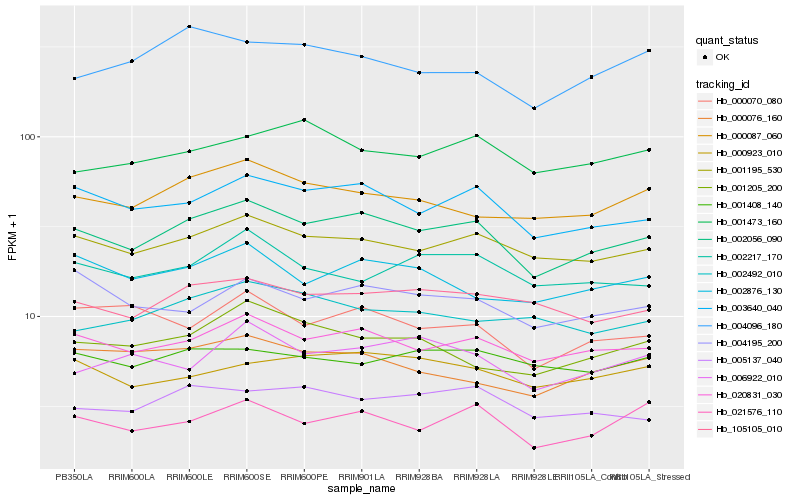
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002056_090 |
0.0 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 2 |
Hb_020831_030 |
0.0495104279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003640_040 |
0.0498253537 |
- |
- |
PREDICTED: uncharacterized protein LOC105647348 [Jatropha curcas] |
| 4 |
Hb_001195_530 |
0.0566061732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_105105_010 |
0.0720113672 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 6 |
Hb_000087_060 |
0.0734434344 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 7 |
Hb_005137_040 |
0.07618752 |
- |
- |
hypothetical protein JCGZ_12324 [Jatropha curcas] |
| 8 |
Hb_021576_110 |
0.0771791568 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 [Jatropha curcas] |
| 9 |
Hb_004195_200 |
0.0790221858 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 10 |
Hb_000923_010 |
0.0792563975 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002876_130 |
0.0793512768 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 12 |
Hb_002217_170 |
0.0813436549 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 13 |
Hb_000070_080 |
0.0818097346 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001205_200 |
0.0836759343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001408_140 |
0.0847131162 |
- |
- |
hypothetical protein JCGZ_00234 [Jatropha curcas] |
| 16 |
Hb_006922_010 |
0.0848591455 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 17 |
Hb_000076_160 |
0.0862362154 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 18 |
Hb_002492_010 |
0.086654315 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 19 |
Hb_004096_180 |
0.0868229514 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 20 |
Hb_001473_160 |
0.0873462385 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |