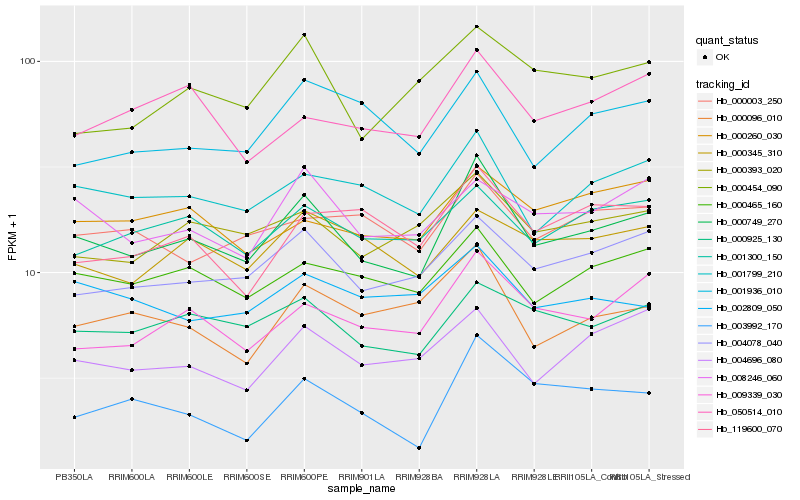
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000749_270 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein 44 [Jatropha curcas] |
| 2 |
Hb_004078_040 |
0.0879877387 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 3 |
Hb_001799_210 |
0.0889391709 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000925_130 |
0.0907537122 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000465_160 |
0.0930551308 |
- |
- |
PREDICTED: uncharacterized protein LOC105632297 [Jatropha curcas] |
| 6 |
Hb_009339_030 |
0.0937411544 |
- |
- |
PREDICTED: mitogen-activated protein kinase 9-like [Jatropha curcas] |
| 7 |
Hb_000393_020 |
0.0985003404 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 8 |
Hb_008246_060 |
0.1018158435 |
- |
- |
hypothetical protein F383_01577 [Gossypium arboreum] |
| 9 |
Hb_001300_150 |
0.1034811931 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 10 |
Hb_004696_080 |
0.1051278017 |
- |
- |
PREDICTED: uncharacterized protein At1g01500-like [Populus euphratica] |
| 11 |
Hb_000345_310 |
0.1056441937 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein isoform X3 [Jatropha curcas] |
| 12 |
Hb_000096_010 |
0.1059137594 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Citrus sinensis] |
| 13 |
Hb_000260_030 |
0.1084134413 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 14 |
Hb_000003_250 |
0.1091947248 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000454_090 |
0.1092679608 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Tarenaya hassleriana] |
| 16 |
Hb_001936_010 |
0.1093158722 |
- |
- |
hypothetical protein Csa_2G348280 [Cucumis sativus] |
| 17 |
Hb_050514_010 |
0.1109825929 |
- |
- |
hypothetical protein 29 [Hevea brasiliensis] |
| 18 |
Hb_003992_170 |
0.1112666674 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 19 |
Hb_119600_070 |
0.1123367727 |
- |
- |
PREDICTED: uncharacterized protein LOC105635190 [Jatropha curcas] |
| 20 |
Hb_002809_050 |
0.1124460819 |
- |
- |
PREDICTED: protein FIZZY-RELATED 2 [Jatropha curcas] |