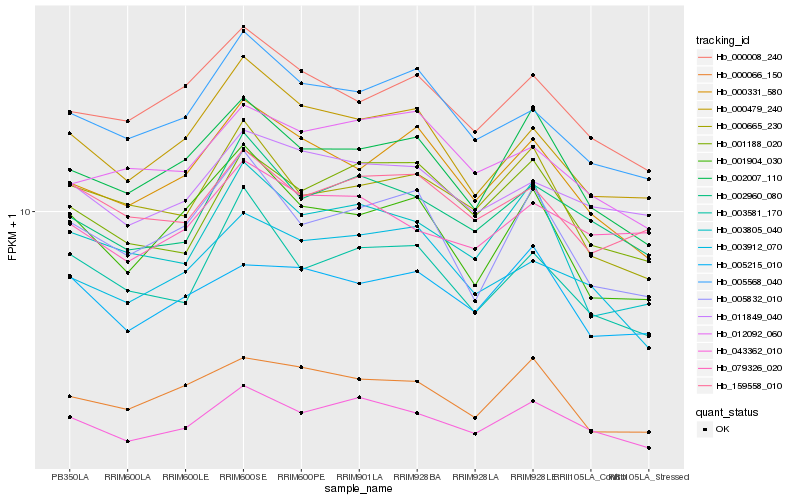
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_043362_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 2 |
Hb_002960_080 |
0.0543693125 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 3 |
Hb_001188_020 |
0.0651560082 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002007_110 |
0.0687755259 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003912_070 |
0.0720006645 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 6 |
Hb_003581_170 |
0.075648611 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003805_040 |
0.0785618789 |
- |
- |
Sacsin [Glycine soja] |
| 8 |
Hb_000479_240 |
0.0803313989 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 9 |
Hb_011849_040 |
0.082437093 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_005832_010 |
0.0839280287 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000331_580 |
0.0840270972 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 12 |
Hb_000665_230 |
0.0848431357 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 13 |
Hb_005568_040 |
0.0862450904 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 14 |
Hb_079326_020 |
0.0883741634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005215_010 |
0.0898376203 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001904_030 |
0.0900249102 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 17 |
Hb_159558_010 |
0.0907652837 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 18 |
Hb_000008_240 |
0.0909586681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_012092_060 |
0.0909766801 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 20 |
Hb_000066_150 |
0.0910195904 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |